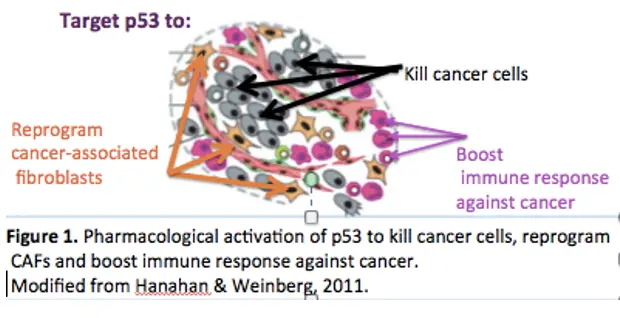
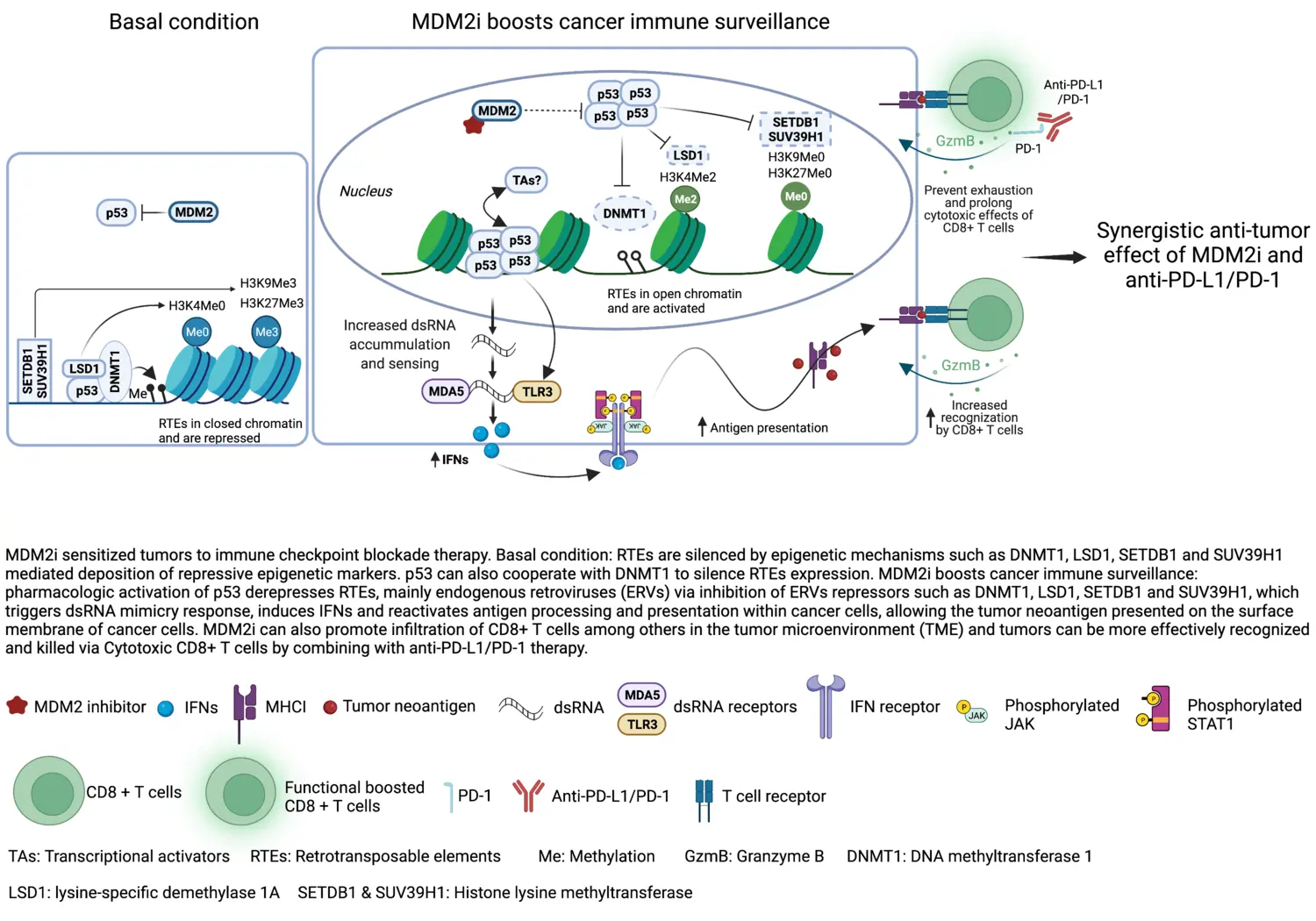
Mechanisms regulating childhood tumors medulloblastoma and neuroblastoma
Medulloblastoma and Neuroblastoma are among the most common malignant neural tumors in children. Neural tumors constitute around one third of all childhood cancers, but almost half of the mortalities. Although advances in therapies have increased survival of the patients, many of the survivors experience complications due to the harsh treatment. This shows not only a need for increasing our understanding of molecular mechanisms operating during neural tumor formation, but furthermore, it highlights the importance of developing targeted therapies that will spare the developing child while specifically eradicating tumor cells.
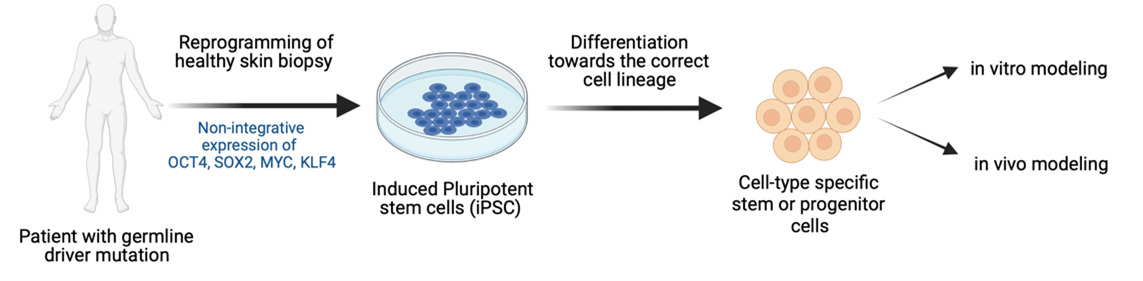
To achieve this, we have developed new cancer models using human disease-relevant cell types. By somatic cell reprogramming to induced pluripotent stem (iPS) cells and differentiation to neural stem cells, we are developing new cancer models with cells from patients with familial driver mutations known to cause medulloblastoma and neuroblastoma.
Identification of novel therapeutic targets
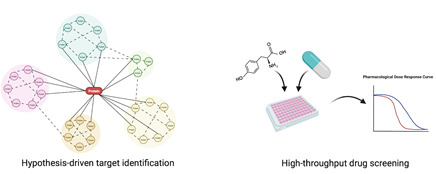
We use our models for identifying molecular mechanisms regulating the disease but also for finding targets that could be used for precision cancer medicine. We combine hypothesis-driven approaches based on mechanistic studies with unbiased exploratory compound library screens to identify novel targets. Targets are validated using our cellular models, in vivo models and in patient material.
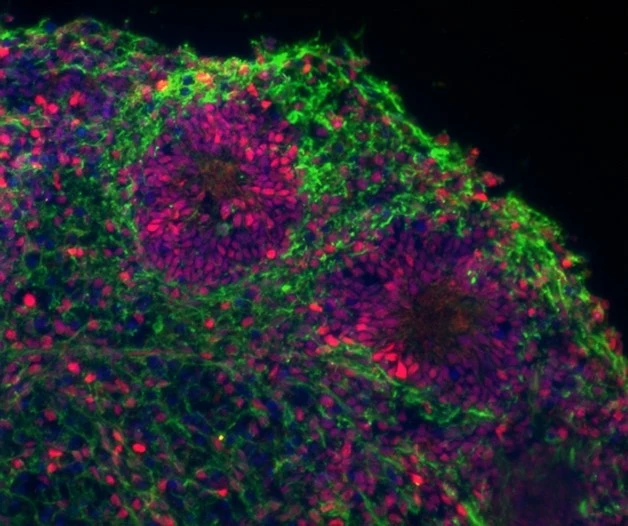
Develop ex-vivo neural organoid models
To create more sophisticated ex-vivo models, we are generating human brain organoids by differentiating iPS cells towards the neural lineage allowing for 3D self-organisation into brain-like structures. This allows us to study how cancer driver-mutations initiate tumor development, how transformed cells interact with neighboring cells, and provides a better model system to evaluate drug responses.
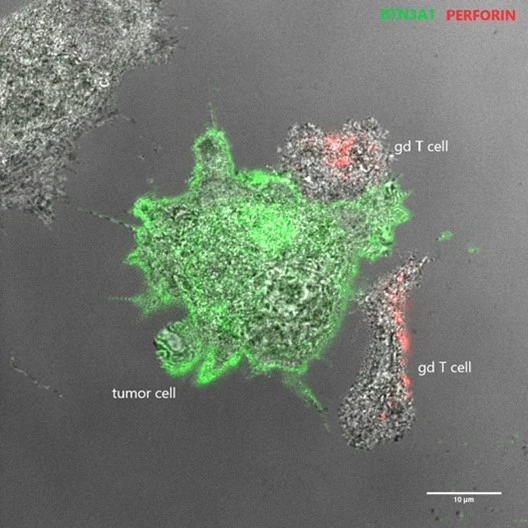
The role of microenvironment in tumor development and how it can be used for cancer therapy
Our lab has a long-standing interest in the cross-talk between tumors cells and the microenvironment, and we have previously studied how tumor angiogenesis, hypoxic responses, drug resistance mechanisms, and immune responses are regulated.
Currently, we are interested how the age of the surrounding environment may affect tumor development. In addition, we are investigating the potential of using non-HLA restricted T cells, also called non-conventional T lymphocytes (NC-T cells), for adoptive cell therapy to eliminate tumors while sparing healthy tissue.
Affiliated research group
Vacancies
The Wilhelm lab are always looking for motivated Postdocs, please apply using contact information below.
Open positions for Doctoral students are posted on KI homepage when available.
Support our research
 Photo: Chokniti Khongchum
Photo: Chokniti KhongchumMake a donation to our research at MTC
Your support means a lot to our success. This allows us to go further in our efforts to improve human health through research and education.
Read here how you can make a donation via Swish.