Our research
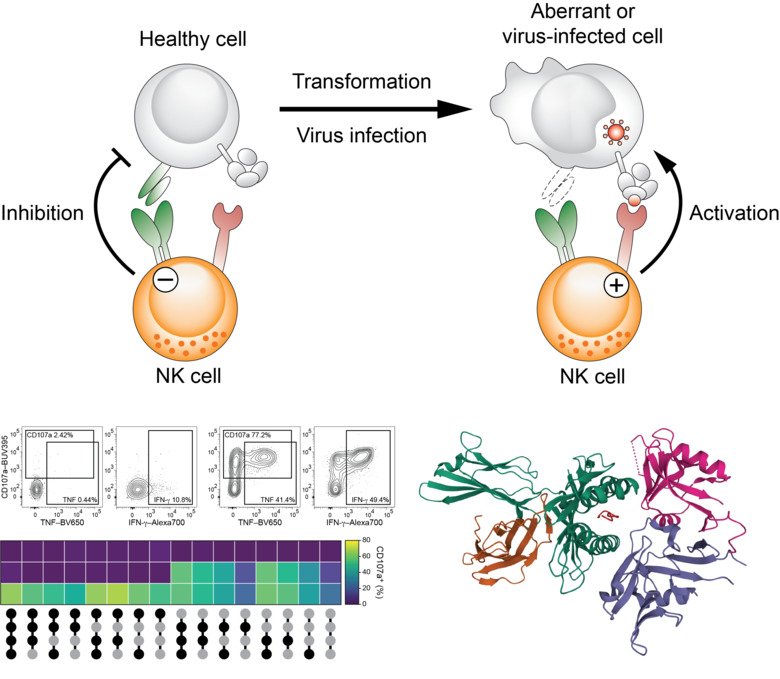
The team explores signals that regulate the activation of immune cells with a focus on cytotoxic effector cells of the innate immune system as well as innate-like cells of the adaptive immune systems. Innate and innate-like lymphocytes with cytolytic properties such as natural killer (NK) cells and unconventional T cells represent the first line of defense against virus infections and developing cancers. Innate cells respond to a broad range of viruses and tumor cells using partly overlapping and partly very distinct pathways.
Central aspects of our research are to delineate the molecular signals that allow innate and innate-like lymphocytes to recognize virus-infected cells and to study by which means these innate cells discriminate between healthy and tumor-transformed cells. For this, we employ a broad range of in silico, cellular, as well as molecular biology techniques and study samples from patients with ongoing disease. In parallel to understanding the signals that initiate and restrict innate immune cell activation, we aim at exploiting such recognition pathways for cell therapy against cancer.
Collaborations
- Professor Chiara Romagnani, German Rheumatism Reserach Center, Berlin, Germany.
- Assistant Professor Amir Horowitz, Icahn School of Medicine at Mount Sinai, New York City, United States of America.
- Associate Professor Mattia Mori, University of Siena, Siena, Italy.
- Assistant Professor Francesco Spallotta, Institute for Systems Analysis and Computer Science “A. Ruberti”, Rome, Italy.
- Professor Martin Messerle, Hannover Medical School, Hannover, Germany.
- Professor Karl-Johan Malmberg, Karolinska Institutet, Stockholm, Sweden.
Research support
- Marie Skłodowska-Curie Actions (EU)
- Hedlunds Stiftelse
- Groschinsky Minnesfond
- Karolinska Institutet Foundation for Virus Reserach
- Region Stockholm
- Stiftelsen Lars Hiertas Minne
- Petrus och Augusta Hedlunds Stiftelse
- Stiftelsen Tornspiran
- KI Foundations (KI stiftelser och fonder)
- Åke Wibergs Stiftelse
- Jonas Söderquist Scholarship

