Probing and Engineering Extrinsically Disordered Proteins to Decode Neuroplasticity
In the Dagliyan Group, we are dedicated to unraveling one of the most profound mysteries of life: How do proteins, through their “extrinsic disorder”, which is the extrinsic modulation of order-disorder transitions, orchestrate cellular processes, including those that govern neuroplasticity, learning, and memory consolidation?

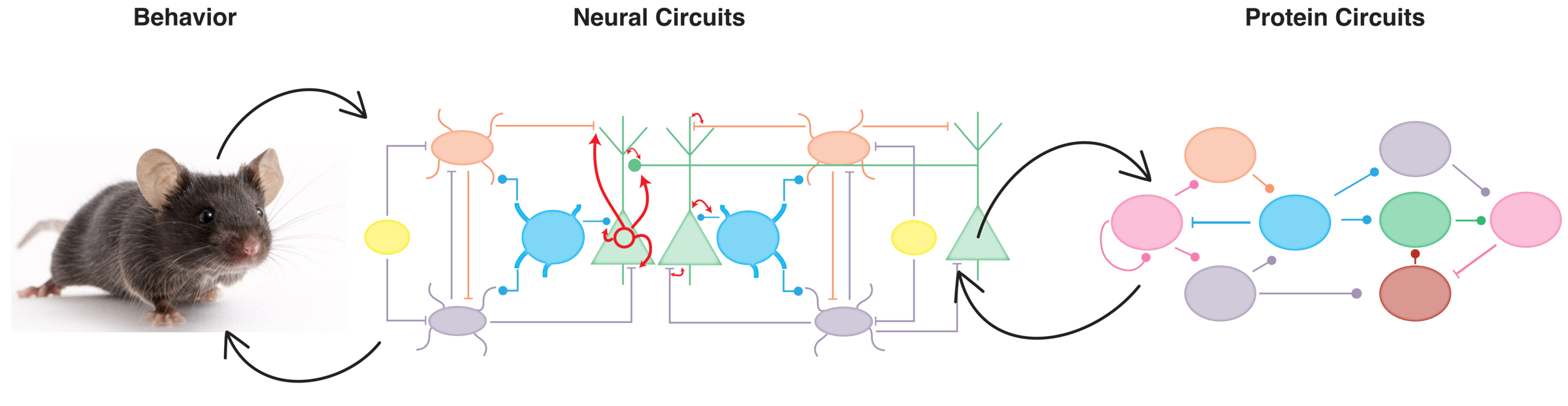
To address this question, we employ cutting-edge technologies to explore protein dynamics with unprecedented precision. We engineer proteins to manipulate and visualize molecular activity in the brains of living mice, delve into the transcriptomic and epigenomic landscapes of individual neurons to uncover the molecular signatures of plasticity, and utilize AI-driven tools to decode complex mouse behaviors, linking them to underlying neural processes. Through advanced recording techniques, we capture neuronal activity both ex vivo and in vivo, offering a window into the brain's action.
Our ultimate goal is to decipher the molecular codes by which neurons translate dynamic protein interactions into functional and behavioral plasticity, using mouse models as a foundational system. By bridging molecular biology and systems neuroscience, we aim to uncover fundamental principles of brain function and pave the way for novel treatments for neurological and psychiatric disorders.
Probing Extrinsic Disorder
Proteins are the workhorses of life, and their ability to switch between states through extrinsic disorder is a fundamental driver of biological processes. Our lab focuses on the extrinsic disorder of proteins and its role in cellular function. Recently, we discovered mechanisms that mediate the disorder-order transitions of proteins, regulating their organization and function in neurons (Dagliyan et al., 2025). Disrupting these transitions impairs neuronal function and behavior, underscoring their critical importance.
Tracing order-disorder transitions of proteins in nuclei.
Engineering Extrinsic Disorder
We also engineer extrinsic disorder to achieve precise control over protein function. One application of this approach is in the study of cell motility, a process that, like many others, is tightly coordinated in space and time. A network of proteins, including GTPases, kinases, GEFs, and others, dynamically orchestrates this process. By designing engineered, extrinsically disordered versions of these proteins, we have successfully manipulated information flow within this network, thereby controlling the motility of fibroblasts, cancer cells, neurons, and neutrophils. This has been achieved through allosteric control of extrinsic disorder using drug- or light-responsive proteins ((Dagliyan et al., 2013; Dagliyan et al., 2016; Dagliyan et al., 2017; Dagliyan et al., 2018; Dagliyan et al., 2019). Below are some examples of engineered extrinsically disordered proteins.
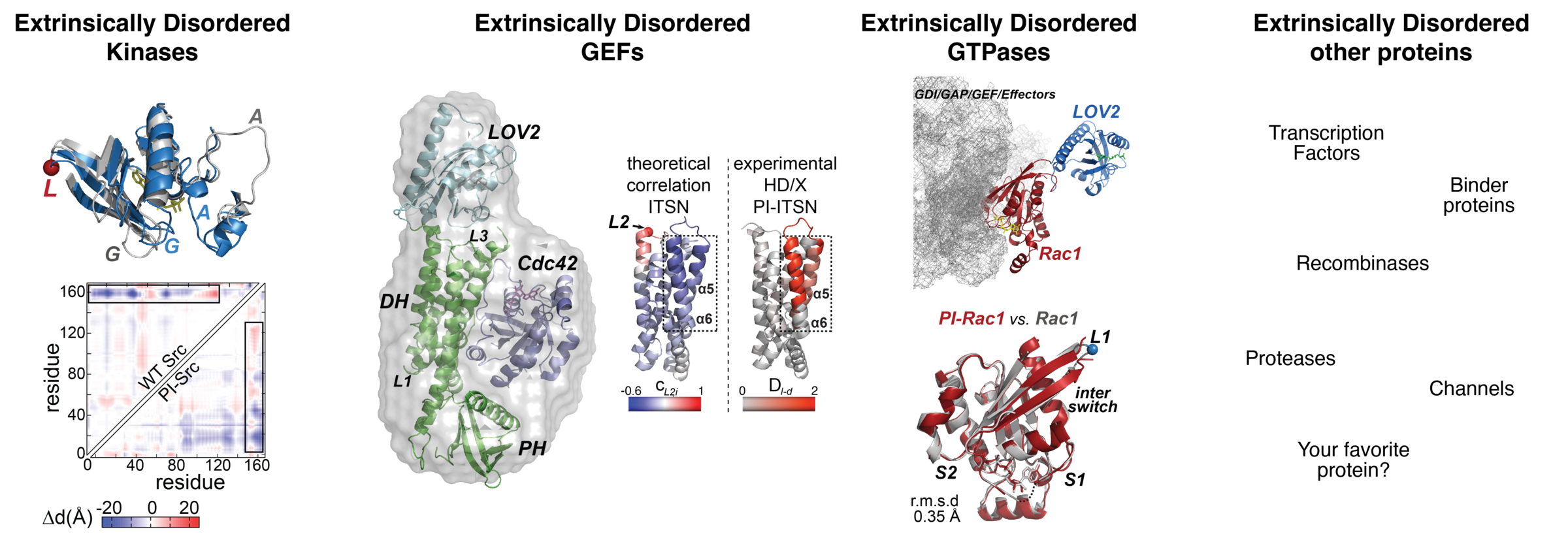
A fibroblast expressing engineered extrinsically disordered Src kinase and its movement is controlled with light.
Fibroblasts expressing engineered extrinsically disordered GEF Vav2 and cellular movement is controlled with light.
A further application involved the design of an extrinsically disordered kinase (LIMK1), which allowed us to precisely manipulate the molecular mechanisms underlying memory formation (Ripoli and Dagliyan, 2023). This engineered kinase, with tunable order-disorder transitions, functioned as a molecular switch that could be activated or deactivated on demand. The results were remarkable: we enhanced memory formation in mice, offering groundbreaking insights into the molecular basis of neuroplasticity.
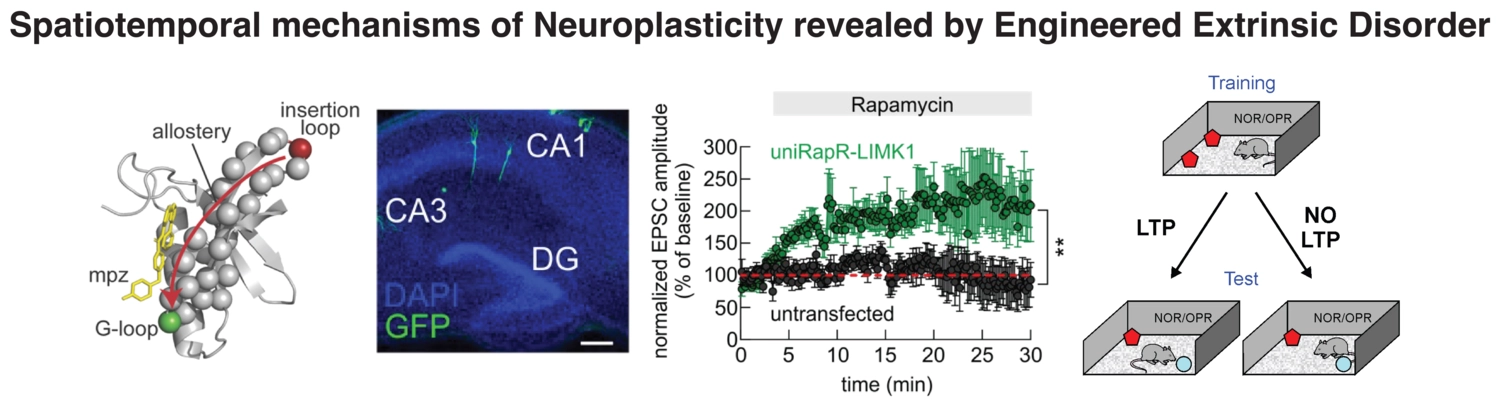
By harnessing the dynamic nature of protein disorder—whether in fully disordered proteins or localized regions of structured proteins—we can design proteins with novel functions, opening new avenues for treating neurological disorders and enhancing cognitive function. In our lab, we combine AI-mediated quantification of mouse behavior, two-photon microscopy for imaging, and in vivo manipulation of protein activity in the mouse brain.
DeepLabCut mediated tracing of mouse behavior.
The Future of Extrinsic Disorder Research
As we continue to unravel the mysteries of extrinsic disorder and refine our ability to engineer it, we aim to apply these principles to uncover new biological mechanisms and develop transformative technologies for human health. We have demonstrated that protein disorder-order transitions are essential for a wide range of cellular processes, from synaptic transmission and cell motility to protein condensation-mediated gene expression. The possibilities for future discoveries are limitless.
We are eager to collaborate with like-minded researchers who share our passion for understanding protein disorder-order dynamics and leveraging these insights to decode the language of nature and engineer life itself.

