We focus on two main areas of research; Primary immunodeficiency and B cell malignancy.
These are our four projects:
- Regulation of immunoglobulin class switch recombination in human B cells
- Induced pluripotent stems cells a platform for personalized diagnosis and therapy in patients with primary immunodeficiency
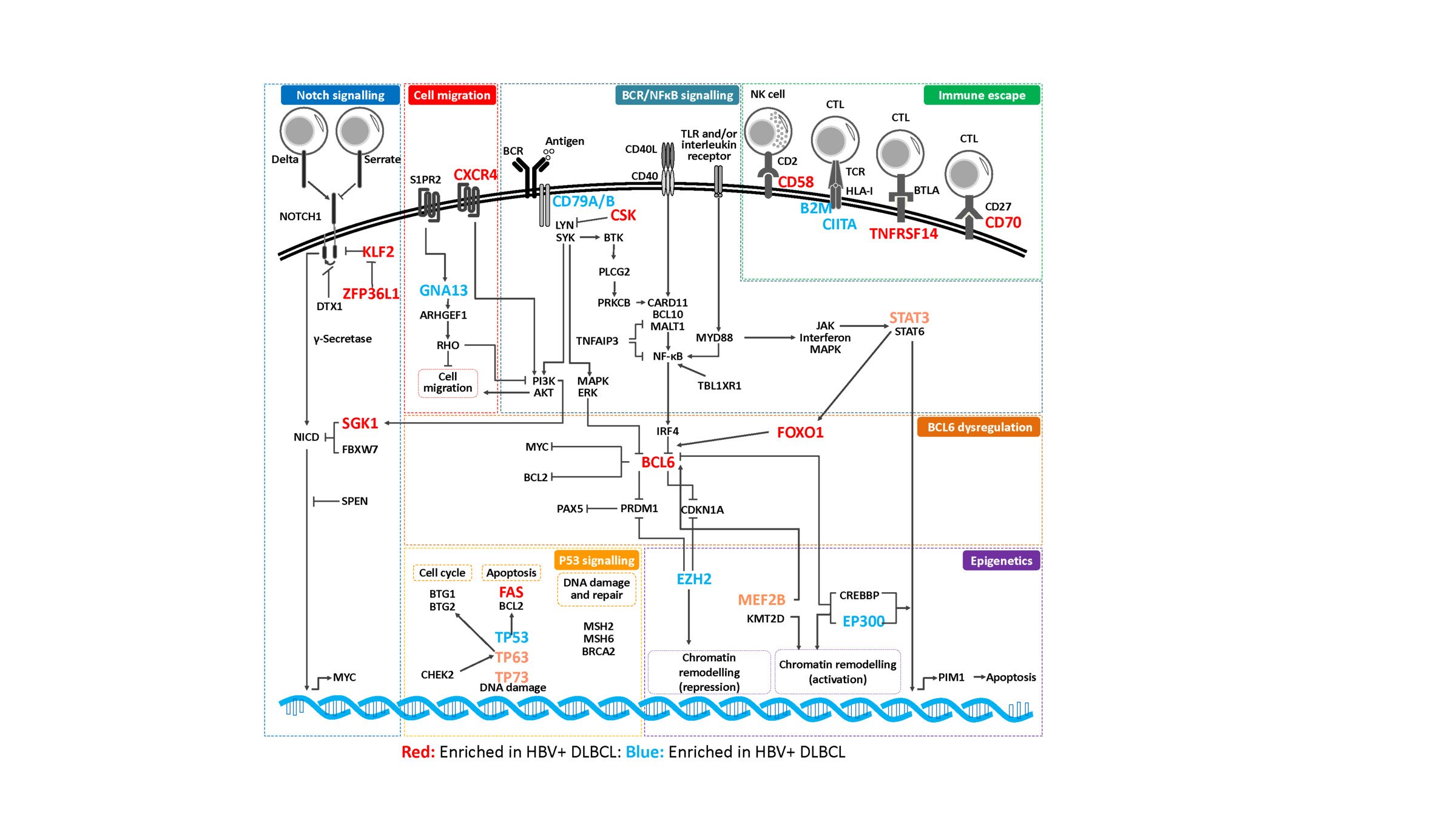
- Discovery of therapeutic targets in B cell lymphoma by next generation sequencing
- Antibody therapy against COVID-19
Research techniques
- DNA sequencing, including whole exome and whole genome sequencing
- Gene expression analysis including real time PCR and RNAseq/transcriptome analysis
- B and T cell functional assays
- Generating iPSCs
- Gene editing using the CRISPR/Cas9 system
Looking for a postdoc position?
Postdoc positions on B cell development, immunogenetics, COVID19 and cancer genetics available.
For details contact: Prof. Pan-Hammarström