
- >
- >
- Perpetration Prevention – Rahm and Joleby's research group
- SOLIID - Sustainable Organizational Learning, Innovation, Improvement and Development in Health and Social services – Monica Nyström's group
- Social Gerontology – Carin Lennartsson group
- Genetic and molecular basis of nervous system disorders – Andrea Carmine Belin group
- Chronobiology – Petrus lab
- Leukemic stem cells – Petter Woll group
- Nanomedicine and Spatial Biology – Teixeira Lab
- Global Disaster Medicine - Health Needs and Response – Johan von Schreeb's research group
- Translational cardiac and skeletal muscle physiology – Daniel Andersson's research group
- Neuroradiology – Staffan Holmin's research group
- Tumour genomics – Lauri Aaltonen's research group
- Neurobiology of Motor Actions – Abdel El Manira group
- Neuroinflammation in Alzheimer disease – Marianne Schultzberg research group
- Palliative Medicin – Linda Björkhem-Bergman's research group
- Nucleotide metabolism and molecular pharmacology – Sean Rudd's Group
- Cardiovascular Inflammation – Research group Peder Olofsson
- Mesenchymal stem cells – Katarina Le Blanc group
- Luigi De Petris' Team
- Autoimmunity and cancer – Research group Marie Wahren-Herlenius
- Atherothrombosis research – Team Nailin Li
- Precision cancer medicine – Olli Kallioniemi's group
- Research group Stephan Mielke
- Genetics of autoimmune diseases – Team Leonid Padyukov
- Musculoskeletal disorders; planetary health & biopsychosocial approaches – Nina Brodin's research group
- Vascular Functions in Metabolic and CNS Diseases – Ulf Eriksson group
- Sarcoma Genomics – Felix Haglund de Flon's Group
- Team Helene Alexanderson
- Injury and Repair in the Immature Brain – Blomgren group
- Statistical methods for cancer patient survival – Therese Andersson's research group
- Individual differences in cognition – Agneta Herlitz’ research group
- Research groups in health economy, healthcare management and policy
- Research groups in reproductive medicine and gynaecology
- Research groups in structural biology
- Research groups in artificial intelligence
- Work environment, health and productivity – Christina Björklund's research group
- Predictive medicine – Mattias Rantalainen's research group
- Epigenetic regulation of leukemia and normal blood development – Andreas Lennartsson's research group
- Epigenetic mechanisms underlying metabolic-inflammatory diseases – Eckardt Treuter's research group
- The IMPACT research group – Marie Löf
- Stockholm Center for Health Economics (StoCHE) – Emelie Heintz's team
- Towards personalized physiology in Intensive Care – Team Johan Mårtensson
- Ninib Baryawno research group
- Neurohormonal Basis of Physiological and Maladaptive Expressions of Survival Behaviors – Stefanos Stagkourakis group
- Breast cancer precision medicine – Marike Gabrielson's research group
- D-WHIN: Danderyd Wrist and Hand INitiative – Research group
- Research group Kilian Eyerich
- Global infections – Research group Anna Färnert
- Translational Microbiome Research and Pandemic Preparedness – Lars Engstrand group
- EcoMind and biological pathways in cognitive aging – Debora Rizzuto group
- Circuits controlling Action and their Evolution – Sten Grillner group
- Mikael Rydén & Niklas Mejhert lab
- Understand Fundamental Immune Mechanisms to Develop New Treatments for Lung Diseases – Tim Willinger group
- Clinical Brain Imaging Methods – Anna Falk Delgado's research group
- Hyperbaric medicine – Peter Lindholm's research group
- Eye movements and vision – Tony Pansell's research group
- Lung toxicology – Lena Palmberg's research group
- The NeuroCardioMetabol Group – Thomas Nyström & Cesare Patrone
- Environmental and genetic factors influence on neurodevelopment – Sandra Ceccatelli group
- Skin wound healing – Research group Ning Xu Landén
- From development to disease: decoding stem cell programs for CNS repair – Erik Sundström's and Xiaofei Li's research group – Erik Sundström-Xiaofei Li group
- Neurodegenerative diseases and Aging – Daniel Ferreira Research Group
- Deep learning models of cancer mechanisms – Avlant Nilsson's Group
- Clinical Physiology – Marcus Carlsson's research group
- Molly Stevens group
- Molecular Mechanisms of Viral Oncogenesis – Maria G. Masucci's Group
- Björn Reinius group
- Quantitative principles in tumour biology – Jean Hausser's Group
- Bone biology – Sara Windahl Research group
- Research group Volkan Özenci
- Genetics and mechanisms of metals – Karin Broberg's research group
- Immunodeficiency Diseases – Lisa Westerberg Group
- Structural studies of fertilisation and zona pellucida module proteins – Luca Jovine's research group
- Tissue regeneration, focusing on fat cells (adipocytes) and obesity – Kirsty Spalding's Group
- Robert Månsson group
- Developmental and Regenerative Stem Cell Medicine – Lanner Lab
- Suicide and mental health lab – Vladimir Carli and Gergö Hadlaczky's group
- Medical, genetic and molecular decoding of neurodevelopmental disorders – Kristiina Tammimies' research group
- Working hours, Recovery and Safety in working life. – Anna Dahlgren's research group
- Research groups in hematology
- SOMIND: Somatic, Mental, and Neurodevelopmental Conditions – Agnieszka Butwicka's research group
- Research groups in drug abuse and addiction
- Research groups in biomaterials science
- Mikael Adner's research group
- Chemical and functional neuroanatomy of pain and stress systems – Tomas Hökfelt group
- Spider silk biology for biomedical applications – Anna Rising's research group
- Cancer Epidemiology and Causal Inference – Research group Maria Feychting
- Protein biochemistry for development of novel medical treatments – Janne Johansson's research group
- Healthcare financing and organization – Clas Rehnberg's group
- Space and environmental physiology – Rodrigo Fernandez-Gonzalo group
- Novel treatment approaches for neuroblastoma – Malin Wickström Näsman research group
- Cancer immune and gene therapy – Rolf Kiessling's Team
- Pancreas biology – Amanda Andersson Rolf's Group
- CAIR-LAB: Clinical AI Research Laboratory – group
- Atopic dermatitis – Research group Emma Johansson
- Reproductive endocrinology and metabolism – Angelica Lindén Hirschberg's research group
- Genetic basis for B and T cell recognition and function – Gunilla Karlsson Hedestam Group
- Cognitive Aging and Mental Health – Erika Jonsson Laukka Group
- Molecular and circuit neuropharmacology – Gilberto Fisone group
- The role of immunity in shaping adipose tissue development and metabolism – Jurga Laurencikiene group
- Chronic myeloid malignancies – Johanna Ungerstedt group
- Studies of tissue microbiology and immunology – Mattias Svensson group
- Sten Linnarsson group
- Balance, gait, exercise and physical activity in neurological diseases – Franzén Group
- Global Health Pharmacology and Therapeutics (GH-Pharma) – Eleni Aklillu's research group
- Real-world-data in Multiple Sclerosis – Anna Glaser's research group
- Suicidology and transcultural psychiatry – Marie Dahlin's research group
- Experimental and Clinical Neuroendocrinology – Maria Petersson's research group
- Cutting edge translational research of common and rare skin disease – Research group Jakob Wikström
- Alzheimer’s Disease Risk Factors & Sex-Specific Mechanisms – Silvia Maioli's research group
- Frontal lobes and cognitive failure – Wahlund's research group
- Clinical Pharmacology – Research group Rickard Malmström
- Molecular Vascular Medicine – Team Lars Maegdefessel
- Molecular endocrine pathology – Christofer Juhlin's Group
- Presynaptic mechanisms – Lennart Brodin group
- Malignant melanoma – Hildur Helgadottir's Group
- Precision strategies to eradicate heterogeneous tumor cells in pediatric cancer – Kasper Karlsson's Group
- Respiratory and systemic immune responses in human pulmonary viral infection and inflammation – Research group Anna Smed Sörensen
- Genetic toxicology – Kristian Dreij's group
- Identifying new molecular targets for the treatment of motor neuron diseases – Eva Hedlund's research
- Epigenetics - basic mechanisms and disease – Karl Ekwall's research group
- Forensic medicine – Henrik Druid's Group
- Ageing and Health – Karin Modig's research group
- Perception Neuroscience – Johan Lundström's research group
- Computational medicine and bioinformatics – Trung Nghia Vu's research group
- Toxicology – Bertrand Joseph´s Research group
- Etiology, prevention and treatment of eating disorders – Ata Ghaderi's research group
- Research groups in medical ethics and history of science
- Research groups in occupational and environmental health
- Research groups in surgery
- Research groups in biophysics
- Research Group Johan Ärnlöv
- Narrative in health and social care – Research group
- Cellular and molecular mechanisms underlying intervertebral disk formation and Diversity and plasticity of adipose tissue cell niche – Meng Xie's research team
- Neuroimmunology of cancer – Sébastien Talbot
- Pediatric Systems Immunology – Petter Brodin's research group
- Microvascular oxidative stress in the triad of cardiovascular, metabolic and renal disease – Mattias Carlström group
- Cardio-renal epidemiology – Juan-Jesus Carrero's research group
- Human Translational Genetics – Stefano Romeo Group
- Clinical Pancreatology – Miroslav Vujasinovic research group
- Hemostasis and Thrombosis – Mika Skeppholms research group
- Movement, Activity and Health – Eva Broström's research group
- Multidimensional health in old age – Amaia Calderón-Larrañaga group
- Molecular epidemiology of aging – Sara Hägg's research group
- Long noncoding RNA in the adipocyte – Alastair Kerr Research Group
- Leukemia niche – Hong Qian group
- Disease mechanisms in severe acute bacterial infections – Anna Norrby-Teglund group
- Gastrointestinal pediatric surgery – Tomas Wester's research group
- Developmental biology and regenerative medicine – Igor Adameyko's research group
- Experimental audiology – Barbara Canlon's research group
- Translational Immunology in Neuroinflammation – Olivia Thomas' research group
- Sickness absence, health and living conditions – Emilie Friberg's research group
- Health Systems and Policy – Cecilia Stålsby Lundborg's research group
- Endocrinology and autoimmune diseases – Research group Sophie Bensing
- Health economics of Alzheimer disease and other dementias – Linus Jönsson Group
- Epidemiology: Diet, contaminants and health – Agneta Åkesson's research group
- Neuronal circuits of anxiety – Janos Fuzik group
- Paediatric rheumatology – Research group Helena Erlandsson Harris
- Cardiovascular Epidemiology – Karin Leander research group
- Molecular cardiology – Research group Francesco Cosentino
- Björn Högberg group
- Regeneration mechanisms – Andras Simon's Group
- Assessing and managing chemical risks – Linda Schenk's research group
- Autoimmunity and adaptive immunity in rheumatic disease – Research group Vivianne Malmström
- Rheumatic systemic diseases – Team Marie Holmqvist
- Heart failure with reduced and preserved ejection fraction. Clinical and translational aspects. – Research group Lars Lund
- Christoph Ziegenhain Group
- Psychiatric epidemiology – Paul Lichtenstein's research group
- Structural basis of RNA biogenesis – Martin Hällberg's research group
- Hematopoietic Stem Cell Biology Group – Sten Eirik W. Jacobsen Lab
- Psychiatric Genetic Epidemiology – Sarah Bergen's research group
- Renal Medicine at Novum (Baxter Novum) – Bengt Lindholm Group
- Research groups in biochemistry
- Psychological treatment: internet and exposure – Brjánn Ljótsson's research group
- Research groups in immunology
- Research groups in ophthalmology
- Research groups in urology and nephrology
- Research groups in sociology and migration
- Integrative physiology – Juleen Zierath and Anna Krook
- Hormonal signalling and cancer – Cecilia Williams research group
- Cell Biology of Cancer – Staffan Strömblad's research group
- Chronic inflammation with focus on rheumatology and cancer – Research group Per-Johan Jakobsson
- Health services and social work research – Lena Von Koch's research group
- Nitrate-nitrite-NO pathway in health and disease – Eddie Weitzberg's research group
- Medical sensors and diagnostics – Team Onur Parlak
- Uncovering the molecular and physical principles governing early embryonic division and nuclear organization – Jan Ellenberg Group
- Nanoscale lipid flux architecture – Veijo Salo’s Research Group
- Residual conditions after polio and neuromuscular diseases – Eva Melin's research group
- Amyotrophic Lateral Sclerosis - ALS – Caroline Ingre's research group
- Saving the brain in critically ill neonates – Ulrika Ådén's research group
- Regulatory Transcriptomics – Kutter group
- NeuroimAging – Grégoria Kalpouzos group
- Section of Pharmacogenetics – MIS-lab
- Biological and clinical aspects of the myelodysplastic syndrome – Magnus Tobiasson group
- Human innate lymphoid cell lab – Jenny Mjösberg group
- Viruses and their interactions with the immune system – Sara Gredmark Russ group
- Protein degradation pathways – Helin Norberg's research group
- Translational Auditory Neuroscience – Christopher Cederroth's research group
- Neuroradiology: Neurodegeneration and Neuroinflammation – Tobias Granberg's research group
- Tumor Immunology and Immunotherapy Group – Dhifaf Sarhan
- Causal inference in epidemiologic research – Arvid Sjölander's research group
- The role of autophagy in Aβ metabolism and neurodegeneration in Alzheimer’s disease – Per Nilsson group
- Darreh-Shori's Lab/group
- PROCOME – Hanna Öfverström's and Marta Roczniewska's group
- Molecular brain imaging of neurodegenerative disorders – Andrea Varrone's research group
- Alcohol and Drug Epidemiology – Mats Ramstedt's research group
- Anxiety related disoders and genetics – Christian Rück's research group
- Psychotherapy Research – Tobias Lundgren's research group
- Human olfaction – Mats J. Olsson's research group
- Tissue stem cells and aging – Pekka Katajisto's Group
- SOSVASC – David Lindström's research group
- Yenan Bryceson group
- Epidemiology of sarcoidosis and systemic lupus erythematosus – Team Elizabeth Arkema
- Viral genomics and metagenomics – Tobias Allander Group
- Defining the metabolic signatures of tissue resident human ILCs – Chris Tibbitt team
- Human papillomavirus which causes cervical cancer – Research Group Karin Sundström
- Research groups in bioinformatics and systems biology
- Cognitive epidemiology – Bo Melin's research group
- Gilad Silberberg group
- Research groups in oto-rhino-laryngology and logopaedics
- Immunoregulation of environmental-induced airway inflammation in severe asthma and bronchiectasis – Apostolos Bossios's research group
- Multimorbidity and frailty in older adults – Davide Liborio Vetrano group
- Psychiatric and pharmaco-epidemiology – Zheng Chang's research group
- Charting normal and malignant hematopoiesis – Team Joakim Dahlin
- Molecular mechanisms governing oxygen homeostasis in development, evolution, and disease – Kragesteen's research group
- Precision Medicine – Infrastructure team
- Research groups in medical epigenetics and epigenomics
- Spinal circuits for whole-body motor coordination – Laurence Picton group
- Clinical HIV epidemiology and comorbidity – Christina Carlander Research group
- Head and neck pathology – Anders Näsman's Team
- Carin Håkansta's research group – Digital work environment and health
- Pediatric and respiratory epidemiology – Catarina Almqvist Malmros' research group
- Sexual and Reproductive Health – Marie Klingberg-Allvin research group
- Angiogenesis, Cancer, Metabolic disease, Cardiovascular disease, Eye disease – Yihai Cao Group
- Cardiovascular and psychosocial health in resilience and healthy brain aging – Chengxuan Qiu Group
- Translational Psychiatry – Catharina Lavebratt's research group
- Rare metabolic liver diseases, coagulation and hepatocellular carcinoma – Stål and Wahlin research group
- Blood Engineering – Vanessa Lundin group
- Development, regulation and function of human innate lymphoid cells throughout life – Jakob Michaëlsson group
- Clinical Genetics – Richard Rosenquist Brandell's research group
- Oxysterols – Ingemar Björkhem and Ulf Diczfalusy research
- Molecular exercise physiology – Carl Johan Sundberg's research group
- Large-scale Network Connectivity in the Human Brain – Peter Fransson's research group
- Environmental physiology
- The Systems Virology Lab – Ujjwal Neogi
- Cancer Cell Invasion – Marco Gerling's research group
- Molecular brain imaging, neuropsychopharmacology, depression and anxiety disorders – Johan Lundberg's research group
- Organization and operation of postural neuronal networks in health and disease – Tatiana Deliagina group
- Paediatrics – Anna Lindholm Olinder's research group
- Petzold group
- Molecular and epidemiological studies of ascending aortic aneurysms – Research group Hanna Björck
- Prevention, Intervention and Mechanisms in Public Health (PRIME Health) – Cecilia Magnusson's research group
- The interplay between circadian 3D genome organization and metabolism in complex diseases – Anita Göndör's Group
- Reconstructive Plastic Surgery and Global Surgery – Jenny Löfgren's research group
- Real-world effectiveness of psychopharmacological treatment – Jari Tiihonen's Research Group
- Center for Heart Failure and Arrhythmia – Gianluigi Savarese
- Sports Medicine – Anders Stålman's research group
- Women's Mental Health Epidemiology – Donghao Lu Lab
- The SMC complexes, chromosome dynamics and stability – Camilla Björkegren's Group
- Extracellular vesicles in immunity – Research group Susanne Gabrielsson
- Precision Cancer Medicine in Lung Cancer - Preclinical, Translational & Clinical Research – Simon Ekman's research group
- RNA-guided DNA repair and cancer – Marianne Farnebo's research group
- Laboratory of translational fertility preservation – Kenny Rodriguez-Wallberg's group
- Research groups in cancer and oncology
- Common Mental Disorders and Behavioral Medicine in Primary Care – Erik Hedman's research group
- Neuronal membrane trafficking – Oleg Shupliakov group
- Research groups in paediatrics
- Biostatistics With Focus on Statistical Methods in Cancer Epidemiology and Screening – Keith Humphreys' research group
- Research groups in nanotechnology
- Gastroenterology – clinical epidemiology and clinical trials – Research group Ola Olén
- Precision engineering of T cells in vivo for therapies of cancer – William Nyberg team
- Protein Misfolding and Prevention by Molecular Chaperones in Cancer and Neurodegenerative Disease – Gefei Chen's research group
- Social Perception – Arvid Guterstam's research group
- Neuroepidemiology – Kyla McKay and Katarina Fink's research group
- Experimental growth research – Lars Sävendahl's research group
- Neuronal circuits of the Basal Ganglia – Maya Ketzef group
- Psychological Interventions - innovation, improvement and implementation – Viktor Kaldo's research group
- Susanna Ranta team
- Neurovascular diseases – Christina Sjöstrand research group
- Lifestyle, Prevention and Health – Ylva Trolle Lagerros research group
- Elias Arnér research group
- Gynecologic Cancer – Henrik Falconer's research group
- B cell biology, from immunodeficiency to cancer – Pan Hammarström Lab
- Developmental Cognitive Neuroscience – Torkel Klingberg group
- Rare Diseases – Ann Nordgren och Anna Lindstrand's research group
- Research on steatotic liver disease – Hagström Group
- Stem Cell Size – Jette Lengefeld Team
- Natural Killer Cell Biology and Cell Therapy – Kalle Malmberg group
- Mast Cell Biology – Research group Gunnar Nilsson
- Traumatic Brain Injuries and Neuromonitoring – Eric Thelin's research group
- Medical Statistics team
- Augmented Neurosurgery – Erik Edström and Adrian Elmi Terander's research group
- Molecular neurophysiology – Karima Chergui's research group
- Injuries Social Aetiology and Consequences (ISAC) – Lucie Laflamme's research group
- Unconventional T cells in human pathology – Andrea Ponzetta team
- Inflammation and immune regulation during infection – Team Christopher Sundling
- Orthopaedics – Anders Enocson's research group
- Endothelial dysfunction in atherosclerotic cardiovascular disease – Research Group John Pernow
- Clinical epidemiology of multiple sclerosis and inflammatory joint diseases – Team Thomas Frisell
- Cancer stem cells and clonal structure in acute lymphoblastic leukemia – Martin Enge's Group
- Mitochondrial metabolism in health and disease – Anna Wredenberg group
- Biomolecular Medicine and Advanced therapies- focus on delivery of RNA therapeutics – Research group Samir EL Andaloussi
- Antimicrobial resistance – Christian Giske research group
- Tissue immunology – Research group Helen Kaipe
- Health Economics and Economic Evaluation – Niklas Zethraeus' group
- Mental Health Neural Dynamics Lab – Kristoffer Månsson's research group
- Stem Cell Biology – Jonas Muhr's Group
- VIVAC research group: Vaccines and Immunotherapies against Viruses And Cancer – Matti Sällberg
- The passage of the mRNP particle through the nuclear pore – Bertil Daneholt's research
- Therapeutic Immunology and Transfusion Medicine – Michael Uhlin’s research group
- Musculoskeletal conditions and sports medicine research – Iben Axén's research group
- Research groups in cancer and oncology
- Nutritional neuroscience – Janina Seubert's Research Group
- Tobias Karlsson's group
- Research groups in pharmacology and toxicology
- Precision Psychiatry – Lu Yi's research group
- Research groups in medicinal chemistry
- Agneta Richter-Dahlfors group
- Cancer epidemiology with focus on breast cancer and applied biostatistics – Anna Johansson's research group
- Autism, ADHD and other neurodevelopmental conditions – Sven Bölte's research group
- Twinstudies on health conditions and sickness absence – Pia Svedberg's research group
- Psychiatric comorbidity and intergenerational transmission of mental health problems – Erik Pettersson's research group
- Genetic and pharmacological epidemiology – Zeberg laboratory
- REACH – digital treatment and prevention – David Moulaee Conradsson's research group
- Mucosal immunology lab – Charlotte Thålin research group
- Stroke – Annika Lundströms research group
- Loh
- Gonçalo Castelo-Branco Group
- Pediatric Neuroimmunology and Epilepsy – Ronny Wickström's research group
- Explores various cell signaling phenomena and their impact on critical biological and medical processes using advanced light microscopy – Per Uhlén Group
- Neural stem cells – Anna Falk group
- Diabetes and Associated Complications – Sergiu-Bogdan Catrina's research group
- Myelodysplastic syndromes – Eva Hellström Lindberg group
- Immune responses to human viral infections and cancer – Hans-Gustaf Ljunggren group
- Health services research in neurological conditions – Charlotte Ytterberg's research group
- Neurosurgery – Mikael A. Svensson's research group
- Westman neuroimaging group
- Stemcells and Inflammation – Lou Brundin's research group
- Pharmacological nitric oxide research – Jon Lundberg's research group
- Epidemiology of Psychiatric Conditions, Substance use and Social Environment (EPiCSS) – Emilie Agardh's & Renee Gardner's Research Group
- Autoimmune neurology – Jakob Theorell team
- Reducing antibiotic resistance – Research group Pontus Nauclér
- Orthopaedics – Karl Eriksson's research group
- Endocrine Surgery – Robert Bränström's research group
- Paediatrics – Inger Kull's research team
- Cancer proteogenomics - from methods to clinical applications – Janne Lehtiö's Group
- Primary Immunodeficiency, Innate Immunity and Antimicrobial peptides – Peter Bergman Research Group/The AMP-group
- Clinical Virology and Immunology – Annika Karlsson's research group
- Jussi Taipale Group
- Global Child Health and the Sustainable Development Goals – Tobias Alfvén's research group
- Topoisomerases, chromatin biology and cancer – Laura Baranello's group
- Stem cells and neural development – Johan Ericson's Group
- Vascular morphogenesis and function in health and disease – Lars Jakobsson group
- Epidemiology and Public Health Intervention Research (EPHIR) – Jette Möller's research group
- Epithelial stem cells in development and disease – Maria Genander's group
- Research Group Gustaf Edgren
- Drug discovery, pancreatic beta-cell regeneration – Olov Andersson's Group
- Unit of Biostatistics – Matteo Bottai
- Research groups in cardiology and cardiovascular diseases
- Sleep, cognition and health – John Axelsson's research group
- Research groups in infectious medicine
- Research groups in physiology and anatomy
- Cardiometabolic epidemiology and aging – Ida Karlsson's research group
- How the overview of research fields is made
- Precision Cancer Control Group – Martin Eklund's research group
- Liver and monocyte remodelling in non-alcoholic fatty liver disease and cardiovascular disease – Rongrong Fan's research group
- Early childhood development and neurodevelopmental conditions – Terje Falck-Ytter's research group
- Mental health and social integration (Mente) – Ellenor Mittendorfer-Rutz's research group
- Mechanisms of protein aggregation and inhibition – Axel Abelein group
- Accelerating drug discovery using molecular modeling and machine learning – Andreas Luttens' group
- Pediatric Healthcare Science – Cecilia Bartholdson research group
- Digital Strategies for AF Detection and Outcome Prevention – Emma Svennberg Team
- Speech-Language Pathology – Liza Bergström's research group
- Arrhythmia – Johan Engdahl's research group
- Environment, nutrition and health – Anna Bergström's research group
- Genetic and epigenetic factors in asthma and allergy – Cilla Söderhäll's research group
- Cardiovascular epidemiology – Team Bruna Gigante
- Neurobiology of Stress and Treatment Response – Juan Pablo Lopez group
- Vascular Surgery – Ulf Hedin's research group
- Annika Bergquist group
- Acute myeloid leukemia – Sören Lehmann group
- Human tissue-resident NK cells – Niklas Björkström group
- Neuropsychoimmunology – Sophie Erhardt's research group
- Diabetes epidemiology – Sofia Carlsson's research group
- Stroke - Acute Intervention and Secondary Prevention – Niaz Ahmed's research group
- Molecular muscle physiology and pathophysiology – Lanner Lab
- Community nutrition and physical activity (CoNPA) – Liselotte Schäfer Elinder's research group
- Inflammatory bowel disease (IBD) – Research group Eduardo Villablanca
- CRISPR-based drug target discovery in cancer and autoimmunity – Research group Fredrik Wermeling
- Emergency care – Kristian Ängeby's research group
- Clinical Cancer Epidemiology – Research group Karin Ekström Smedby
- Cancer prevention and screening – Johannes Blom's research group
- Somatosensation – Patrik Ernfors group
- DNA replication & Cancer Genetics – Lemmens Group
- Laboratory testing for alcohol and drugs of abuse – Anders Helander research
- Anaesthesia and Intensive care – Rebecka Rubenson Wahlin/Anna Schandl's research group
- Jonas Fuxe Group
- SCF ubiquitin ligases, cell cycle, transcription and cancer development – Olle Sangfelt's Group
- Toxicological Mechanisms – Emma Wincent's research group
- Stem Cells in Tissue Homeostasis and Regenerative Medicine – Jonas Frisén's Group
- Urology – Olof Akre's research group
- The New World of Work – Theo Bodin's research group
- Midbrain dopaminergic neuron development
- Research group Michael Fored
- Neurobiology of pain & Therapeutics – Saida Hadjab Group
- Risk assessment – Mattias Öberg's research group
- Research groups in cell and molecular biology
- Developmental Psychology: Digital Media and ADHD – Lisa Thorell's research group
- Research groups in education and pedagogy
- Research groups in biostatistics and probability theory
- ESSI – Emotion regulation, Self-injury, Suicide, and Intervention – Johan Bjureberg's research group
- Medical ethics – Gert Helgesson's group
- Treatment of substance use disorders – Johan Franck's Research Group
- Sex hormones and sex differences in diseases of the brain – Ivan Nalvarte's research group
- Cilia in the brain – and their connections to human brain disorders – Peter Swoboda's research group
- Evidence-based methods in autism and ADHD – Tatja Hirvikoski's research group
- Long-Term Outcomes after Perioperative and Intensive Care – Max Bell research group
- Translational and clinical research in acute myeloid leukemia – Team Martin Jädersten
- CEREBRA – Susanne Palmcrantz's research group
- Ute Römling group
- Perioperative quality – Jan Jakobssons research group
- Translational Pharmacology – Kent Jardemark's team
- Global and Sexual Health (GloSH) – Anna Mia Ekström's research group
- Medical Inflammation Research – Rikard Holmdahl group
- Neuro-infections & Neuroinflammation – Federico Iovino Group
- Clinical Chemistry and Blood Coagulation – Jovan Antovic's research group
- Translational research on human microbial infections and its consequences – Anders Sönnerborg's group
- Human tissue-resident NK cells in homeostasis and disease – Nicole Marquardt team
- Etiology and pathogenesis of type 1 diabetes – Malin Flodström-Tullberg group
- Oxygen sensing, cancer and intratumor heterogeneity – Schlisio Lab
- Genetic Epidemiology of Neuroinflammatory Disorders – Ingrid Kockum's research group
- Retina – Anders Kvanta's research group
- Glaucoma – Pete Williams' research group
- Leadership in healthcare and academia – Mia von Knorring's group
- Luminal gastroenterology – Research group Charlotte Hedin
- Identifying molecular signals in the genital mucosa that determine susceptibility to sexually transmitted infections – Research group Kristina Broliden
- Coronary heart disease – Research group Per Svensson
- Educational development – Matti Nikkola's group
- Post-transcriptional regulation in mitochondria – Joanna Rorbach group
- Cellular diversity is generated during development, neuronal identity – Johan Holmberg's Group
- Lipid handling in health and cardiometabolic disease – Research group Carolina Hagberg
- Renal glomerulus biology and diseases – Jaakko Patrakka group
- Research group Caroline Palm Apergi
- Precision pathology and tumor heterogeneity – Johan Hartman's Group
- Lifelong Learning in Health Care Contexts – Terese Stenfors' group
- Translational Arthritis Research – Team Bence Réthi
- Stem cells, Developmental Biology, Heart Disease, Heart Development, Genetics, Biotechnology – Kenneth R. Chien's group
- Urological cancer – Lars Egevad's Group
- The ubiquitin/proteasome system in neurodegenerative diseases and cancer – Nico Dantuma's Group
- Experimental alcohol- and drug dependence research – Vladana Vukojević research group
- Chronic inflammatory disease epidemiology – Research group Johan Askling
- Statistical and bioinformatics analyses of high-throughput molecular data – Yudi Pawitan's research group
- Urban environment and children´s and youth health – Olena Gruzieva's research group
- Research groups in laboratory medicine
- Research groups in medical biotechnology
- Research groups in physiotherapy
- Breast cancer epidemiology – Kamila Czene's research group
- Research groups in human computer interaction
- Research group Liv Eidsmo
- Preventive Medicine – Susanna Larsson's research group
- Transition to adulthood for individuals with neurodevelopmental conditions – Ulf Jonsson's team
- Function and Health in Respiratory and Cardiovascular Conditions – Malin Nygren-Bonnier's research group
- Laboratory of lymphocyte biology – Team Taras Kreslavskiy
- Global Reproductive Outreach & Wellness (GROW) – Michael Wells
- Mats Marshall Heyman research group
- SLE/APS/Vasculitis group – Research group Aleksandra Antovic
- Itziar Martinez Gonzalez group
- CoNFIND Cognition and Fatigue – Marika Möllers research group
- Tissue immunosurveillance by cytotoxic lymphocytes – Takuya Sekine team
- Physical activity and Sports medicine with focus on prevention – Hagströmer research group
- Receptor biology and signaling – Schulte Lab
- Neurobiology of Obesity – Alessandro Furlan group
- Upper GI Surgery – Jesper Lagergren's research group
- Sepsis and COVID-19 – Kristoffer Strålin Group
- Harnessing Neutrophils for Precision Cell Therapy against solid tumors – Roland Fiskesund Team
- Human T cell immunity to evolving viruses and cancers – Marcus Buggert group
- Neuroradiology - MRI physics – Stefan Skare's research group
- Wilhelm lab
- Inflammatory responses in cancer and autoimmunity – Mikael Karlsson's Group
- Cultural Medicine – Solvig Ekblad's unit
- Working life, ergonomics, psychosocial factors, and health – Research group Daniel Falkstedt
- Aging and health – Welmer's research group
- NK Cell Recognition – Klas Kärre Group
- Computational Breast Imaging – Fredrik Strand's Group
- Drug treatment – Erik Eliasson research group
- Neural differentiation as a strategy for neuroblastoma treatment – Marie Arsenian Henriksson Group
- Cancer immunotherapy – Mattias Carlsten group
- Cell-based immune therapy for cancer – Andreas Lundqvist's Group
- Basic molecular structure of human skin – Lars Norlén's research
- Reproductive, Perinatal and Pediatric Epidemiology – Research group Olof Stephansson
- Research group Erik Melén
- Research group Joel Nordin
- MINT – Klas Karlgren's team
- Translational breast cancer research – Theodoros Foukakis' Group
- Genes function and molecular bases of diseases – Research group Magdalena Paolino
- Homeostasis and tissue repair mechanisms in the mammalian system, pericytes, stem cells – Christian Göritz Group
- The Perinatal Epidemiology Lifecourse Lab – Team Neda Razaz
- Experimental Cancer Medicine (ECM)
- Research group Therese Djärv
- Molecular and cellular neuroendocrinology – Tibor Harkany group
- Neuropsychiatric disorders – Håkan Karlsson group
- Research groups in dermatology and venereal diseases
- Evidence-based practice: prevention, intervention and implementation – Pia Enebrink's research group
- Research groups in radiology and medical imaging
- Research groups in psychiatry
- Maria Eriksdotter research group
- Research groups in precision medicine
- Breast cancer surgery – Jana de Boniface's research group
- ICare – Ann Langius-Eklöf's research group
- Immunology and Chronic Disease – Johan Frostegård's research group
- Health in Everyday Life (HELD) – Susanne Guidetti's research group
- Center for Resuscitation Science – Jacob Hollenberg's research group
- Applied Developmental Neurobiology – Carl Sellgren research group
- Reproductive Health/Reproductive Medicine – Kristina Gemzell Danielsson's research group
- Malin Holzmann's research group
- Radiation therapy/ Radiation Oncology – Pehr Lind Group
- Fossati
- APP processing and Abeta localization at super-resolution and glycan biomarkers for Alzheimer disease – Sophia Schedin Weiss's research group
- ALGOSH: Algorithmic Management at Work
- Development of autonomic control – Herlenius Research
- Maintenance and expression of mtDNA in disease and ageing – Nils-Göran Larsson Group
- Neuropharmacology - Movement Disorders – Per Svenningsson's research group
- Somatosensation & Gargalesis – Konstantina Kilteni group
- Signal Transduction – Ismael Valladolid Acebes' research group
- Microbiome and Infections – Piotr Nowak Research Group
- Immunological tolerance and transfusion immunology – Petter Höglund group
- Immunopathogenesis and new therapeutic strategies in tuberculosis – Susanna Brighenti group
- Pain and Brain Imaging – Karin Jensen's research group
- Susanne Nylén group
- Hand surgery – Maria Wilckes Research group
- Environmental impact on pulmonary host defence and chronic airflow obstruction – Anders Lindén's research group
- Rehabilitation, collaboration and aging – Elisabeth Rydwik's research group
- Tuberculosis Aerobiology – Antonio Rothfuchs group
- Oscar Fernandez-Capetillo group
- Diversity within the T cell response, T cell memory – Research group Carmen Gerlach
- Clinical and translational melanoma research – Hanna Eriksson's Group
- Meiotic chromosome segregation – Christer Höög's research group
- B cells and autoantibodies in rheumatic disease – Team Caroline Grönwall
- Precision medicine in lymphoid malignancies including CLL; from new targets to real-world assessments – Anders Österborg's Group
- Research group for Cancer evolution
- Jiri Bartek group
- Translational Cardiology (@TransCardio) – Research group Magnus Bäck
- Gene regulation, genome-wide experimental and computational techniques – Rickard Sandberg's Group
- The Helleday Laboratory focuses on harnessing defects in the DNA damage response and metabolism to develop novel therapies
- Small RNAs in cancer development – Weng-Onn Lui's Team
- Geriatric pharmacoepidemiology – Kristina Johnell's research group
- Molecular neurodevelopment and neuro-oncology – Ola Hermanson group
- Research groups in anesthesiology and intensive care
- Gastrointestinal epidemiology – Jonas Ludvigsson's research group
- Health inequalities and minority stress – Richard Bränström's research group
- Research groups in microbiology
- Research groups in orthopaedics
- Hereditary hematological malignancies – Bianca Tesi team
- Lars Holmgren's Group
- Substance use prevention – Johanna Gripenberg's research group
- Perioperative care – Ulrica Nilsson's research group
- Mechanisms behind HIV-1 latency and rebound – Peter Svensson's group
- ENGAGE- Enacting health and change through everyday activity – Patomella group
- Diagnosis and treatment of children with asthma and allergy: from molecular diagnosis to intervention – Jon Konradsen's research group
- Translational research in childhood cancer and histiocytic diseases – Nikolas Herold Research group
- Coronary artery disease – research at Danderyd hospital
- Mechanisms of malignant hematopoiesis – Pedro Moura team
- Regulation of B cell activation – Pia Dosenovic Group
- Complex interventions – Team Carina King
- Reproductive endocrinology and metabolism – Elisabet Stener-Victorin's research group
- Tissue engineering – Magdalena Fossum team
- Inflammation and metabolism – Nicolas Pillon's team
- Neurobiology of Sensory Systems – François Lallemend group
- Experimental Traumatology Research Unit
- Genetic Modification of NK cells for Optimized Functions against Cancer – Arnika Wagner team
- NK cells in the development of adaptive immune responses – Benedict Chambers team
- Molecular pain research – Camilla Svensson's research group
- Brain Connectomics – Joana Pereira's research group
- Genomic Science and RNA Biology – Vicent Pelechano group
- Immunotherapy – Robert Harris' research group
- Infectious diseases "Venhälsan" – Jaran Eriksen's research group
- Musculoskeletal disorders from a biopsychosocial perspective – Wim Grooten's research group
- Hjerling-Leffler group
- Respiratory and invasive infections and microbial pathogenesis – Birgitta Henriques-Normark Laboratory
- Multimodal Brain Imaging – Daniel Lundqvist's research group
- Obsessive - Compulsive and Related Disorders Across the Lifespan – David Mataix-Cols research group
- With a focus on proteins for better cancer treatments – Pär Nordlund's Group
- Precision cancer medicine and precision radiotherapy in lung cancer - from molecular mechanisms/biomarkers to clinical trials – Kristina Viktorsson's team
- Cancer Bioinformatics – Nick Tobin's Team
- Lipoproteins and the Immune System – Research group Stephen Malin
- Equity and Health Policy (EHP) – Ann Liljas' research group
- Effects of hypoxia in physiological and pathological contexts, Hypoxia Inducible Factors (HIF) – Randall Johnson's Group
- Chemistry II – Jesper Z. Haeggström group
- Genomic analysis of parasites and viruses, metagenomic sequencing – Björn Andersson's Group
- Translational control of cancer – Ola Larsson's Group
- Gastrointestinal Surgery KI SÖS – Gabriel Sandblom's research group
- Understanding the mechanisms behind hantavirus-mediated pathogenesis – Jonas Klingström group
- The Gynaecological research team – Elisabeth Epstein
- Understanding how life develops – Emma Andersson's Group
- Statistical methods in epidemiology – Paul Dickman's research group
- Neurons and Neural Networks – Ole Kiehn group
- Research groups in developmental biology
- Behavioral, Resilience and Digital Health in Pain – Rikard Wicksell’s research group
- Research groups in neurology
- Research groups in epidemiology, global public health and social medicine
- Nordic Brain Network – Miia Kivipelto's research group
- Resilience and mental health – Serhiy Dekhtyar group
- Forensic behavioral and neuroscience – Katarina Howner's Research Group
- Immune Engineering – Team Leo Hanke
- Chemical and physical exposure in the work environment and health – Jenny Selander's research group
- Mechanobiology of cardiac regeneration – Elif Eroglu's Group
- Tumors of the female genital organs – Miriam Mints' research group
- Psychoneuroimmunology – Mats Lekander's research group
- Research groups in odontology
- Neuroimmunovascular biology – Harald Lund's team
- Improved synthetic DNA structures by evolutionary selection – Erik Benson Group
- Lifestyle and Kidney Cancer – Stephanie Bonn Team
- Neuroplasticity and Regeneration – Konstantinos Ampatzis group
- Paediatric cell and molecular biology – Hjalmar Brismar's research group
- Quantitative Biology of the Nucleus – Magda Bienko Group
- Cardiometabolic factors, brain aging, and dementia care – Weili Xu group
- Cognitive Neuroscience of Body and Self – Henrik Ehrsson Group
- Surgical Care Science – Pernilla Lagergren's research group
- Clinical and translational studies on viral hepatitis – Soo Aleman group
- Cell and Gene Therapy – Evren Alici group
- Personalized Medicine and Drug Development – Lauschke-lab
- Electrophysiological neuropharmacology – Göran Engberg's research group
- Mechanisms of Pain and Treatment – Eva Kosek's research group
- Functional (Epi)genomics of Neuroinflammatory Diseases – Maja Jagodic's research group
- Molecular basis of gene regulation of diseases – Carsten Daub's research group
- Health risk assessment methodology – Anna Beronius' group
- Medical genetics – Catharina Larsson's Group
- Translational Genetics of Neurodegenerative disease – Caroline Graff's research group
- Development, transcription factors, neurons, dopamine – Thomas Perlmann's Group
- Clinical and translational research within lupus and autoimmunity – Team Ioannis Parodis
- Cell cycle, mitotic entry, and DNA-damage checkpoints – Arne Lindqvist's Group
- Breast Surgery – Irma Fredriksson's research group
- Laura Orellana's Group
- Research group Ali Mirazimi
- Early drug discovery research in chronic inflammatory diseases – Research group Michael Sundström
- Integrative Physiology – Juleen Zierath's research group
- Simon Elsässer group
- Clinical Neuroimmunology and Immunomodulation – Fredrik Piehl's research group
- Understanding mechanisms of vaccination – Research group Karin Loré
- Systems regenerative neurobiology – Enric Llorens Group
- Clinicial Cancer Genomics – Johan Lindberg's research group
- Jeanette Hellgren Kotaleski group
- Brain control of food intake and body weight – Björn Meister group
- Research groups in endocrinology and diabetes
- Research groups in gastroenterology and hepatology
- Research groups in neurosciences
- Research groups in respiratory medicine and allergy
- Caring in Community Care – Zarina Nahar Kabir's research group
- Molecular mapping of the nervous system in health and disease – Jan Mulder group
- Child and adolescent psychiatry – Jens Högström's research group
- Genetic mechanisms of ageing – Maria Eriksson's research group
- Digital Psychiatry – Philip Lindner's research group
- Synaesthesia, autism and perception – Janina Neufeld's team
- Vascular complications in cardiometabolic disease – Team Zhichao Zhou
- Lars Karlsson team
- Sepsis and microcirculation – Sara Tehrani's research group
- Translational Clinical Orthopaedics – Michael Axenhus Research Group
- Antigen-specific T cell repertoires – Cassotta lab
- Translational Molecular Imaging – Nordberg Lab
- Neuroplasticity – Dagliyan group
- Developmental and Translational Neurobiology – Cristiana Cruceanu's research group
- Microbiota–gut–brain axis and neurodevelopment – Rochellys Diaz Heijtz group
- Thoracic Surgery – Anders Franco-Cereceda's research group
- Electrophysiology – Mats Jensen-Urstad group
- Role of T cells in human host defense – Johan Sandberg group
- Germ cell biology and developmental programming in epigenetic inheritance of diseases – Qiaolin Deng's research group
- Unit for Bioentrepreneurship – Hanna Jansson's group
- Health informatics – Sabine Koch's group
- Cancer, rapid ageing and nutrition – Martin Bergö's research group
- Clinical radiation therapy – Åsa Carlsson Tedgren's Group
- TRANslational Theranostics Group – Thuy Tran
- Pathogenic pathways in Alzheimer Disease – Lars Tjernberg's research group
- Hematological and solid tumors – Research group Xu Dawei
- Developmental Neurogenomics – Michael Ratz's Group
- Notch signaling, ES cells, myogenic and vascular progenit breast cancer – Urban Lendahl's Group
- Particle toxicology – Hanna Karlsson's group
- Mitochondrial dysfunction in Alzheimer Disease – Maria Ankarcrona's research group
- Molecular pathology of the lung and pleura – Katalin Dobra's Group
- Late effects and cancer survivorship after diagnosis and treatment of aggressive lymphoma – Team Sandra Eloranta
- Research Group Anna Nopp
- Coding and non-coding RNAs in cancer – Per Hydbring's Team
- Research group Mikael Björnstedt
- Sister chromatid cohesion in DNA damage responses, DSB repair, Genome Integrity, Gene regulation – Lena Ström's Group
- Inborn Errors of Endocrinology and Metabolism – Anna Wedell's research group
- Functional Precision Medicine – Brinton Seashore-Ludlow's Team
- Understand skin using modern biology – Maria Kasper's Group
- Integrative Epidemiology – Fang Fang's research group
- Molecular Biometry – Roman Zubarev group
- Idiopathic pulmonary fibrosis (IPF) – Research group Magnus Sköld
- Molecular Neuroscience – Carlos Ibáñez group
- Research groups in psychology
- Research groups in forensic science
- Research groups in medical genetics and genomics
- Research groups in nursing
- Research groups in rheumatology and autoimmunity
- Genetic epidemiology of prostate and testicular cancer – Fredrik Wiklund's research group
- Systems biology of aging – Juulia Jylhävä's research group
- Aging – basic mechanisms and interventions – Christian Riedel's research group
- Embryonal, foetal and brain development – Juha Kere's research group
- Research groups in occupational therapy
- Growth and Cartilage Biology – Ola Nilsson's research group
- Infections and immunity in children with cancer – Anna Nilsson's research group
- Oxygenation, early resuscitation in trauma and drug treatment in cardiac arrest – Team Malin Jonsson Fagerlund
- Prehospital Emergency Care – Veronica Vicente's Research Group
- The brain after surgery and trauma - neuroinflammation, cognitive impairment and dementia – Lars I Eriksson research group
- Personalized diet and medications for cognition – Garcia-Ptacek group
- Arthroplasty – Olof Sköldenberg’s research group
- Anna Undeman Asarnoj team
- Visual Neuroscience – Rune Brautaset's research group
- Chemicals and female fertility – Pauliina Damdimopoulou Research Group
- Molecular and cellular exercise physiology – The Ruas Lab
- Neural circuits of cognition – Marie Carlén group
- Colorectal Surgery – Anna Martling and Caroline Nordenvall's research group
- Myeloma group – Evren Alici's research group
- Medicine and history of science – Eva Åhrén's group
- Cardiometabolic diseases – Ping Chen research group
- Anders Mutvei research group
- Ocular Oncology and Pathology – Gustav Stålhammar's research group
- Clinical Neurophysiology – Charith Cooray's research group
- Regulation of Gene Expression during Viral Infection – Gerald McInerney Group
- Social and affective learning and decision-making – Andreas Olsson's research group
- Translational Breast Cancer Research: Long-Term Risk and Endocrine Treatment Benefit – Linda Lindström's Group
- Notch3 and the cerebral small vessel disease CADASIL – from molecular mechanisms to treatment strategies – Helena Karlström's research group
- Dementia and Multimorbidity – Dorota Religa's research group
- Diagnostic Radiology – Lennart Nedar's research group
- Cognitive Neuropsychiatry – Predrag Petrovic's research group
- Chemical carcinogenesis – Ulla Stenius' group
- Metal toxicology – Maria Kippler's research group
- Cellular Stress in Health and Disease – Federico Pietrocola's group
- Autophagy and cardiovascular disease – Team Ewa Ehrenborg
- Research group Birgitta Sander & Birger Christensson
- Innate Immune Regulation – Quirin Hammer team
- Genetics of tumor predisposition and progression in breast cancer – Svetlana Bajalica Lagercrantz's Team
- Implementation and Quality (IMPAQT) – Claudia Hanson's research group
- Structural and Biophysical Immunology – Research group Adnane Achour
- Ulrika Marklund group
- Sustainable work & occupational safety and health – Emma Brulin's research group
- Health Systems Leadership, Management, and Safety – Carl Savage's group
- Cell and Molecular Immunology - Research group Cecilia Söderberg Naucler
- Brain Circuits – Konstantinos Meletis group
- Petter Ljungman's research group- Health effects of the Ambient Environment
- Neuropsychology of music – Fredrik Ullén group
- Research groups in geriatrics and gerontology
- Research groups in nutrition and dietetics
- Research groups in sports and fitness sciences
- Prostate Cancer – Henrik Grönberg's research group
- Methods of health promotion in the workplace – Lydia Kwak's research group
- Autism and perinatal epidemiology – Sven Sandin research group
- Wengström's research group
- Modelling and simulation with applications to cancer epidemiology and health economics – Mark Clements' research group
- Health Technology Assessment – Monica Hultcrantz’s group
- Pediatric anesthesiology Stockholm – Per-Arne Lönnqvist's research group
- Biophysics – Erdinc Sezgin's Lab
- War, crisis, and security studies
- Systems Medicine – Johan Björkegren research group
- Trauma – Max Gordon's research group
Section of Pharmacogenetics – MIS-lab
Our research aims at elucidate the bases for interindividual differences between individuals in liver function, risks for liver diseases, drug response and risk for adverse drug reactions as well as to identify new drug targets for treatment of steatosis and liver fibrosis, using a novel in vivo-like 3D liver spheroid system.

Latest news
MIS-lab
Our research aims at understanding the basis for liver diseases, drug induced injuries and the differences between individuals in liver function, risk for liver diseases, liver regeneration, human drug metabolism, drug toxicity and drug response, thereby facilitating a more effective personalized drug treatment at the clinics.
Background
The liver is a vital organ for synthesis and detoxification. It has an ability to regenerate after partial hepatectomy and other major injuries, however the mechanisms behind are relatively unknown.
If major liver disease occurs transplantation is today the only manner for cure. The most significant liver diseases are:
- hepatitis, inflammation of the liver, mainly induced by various viruses but also drugs alcoholic liver disease including fatty liver disease
- alcoholic hepatitis, and cirrhosis, the formation of fibrous tissue in the place of liver cells that have died
- non-alcoholic fatty liver disease (NAFLD), where hepatic steatosis, accumulation of lipids within hepatocytes, may lead to inflammatory disease (i.e. steatohepatitis) and cirrhosis
- non-alcoholic fatty liver steatohepatitis (NASH) and v) cirrhosis, where the developing fibrosis can cause chronic liver failure. NAFLD constitutes a clinicopathological condition that encompasses a spectrum of liver disorders.
NAFLD is the most common liver disease affecting between 20% and 44% of European adults and 43-70% of patients with type 2 diabetes (Blachier et al, 2013). Onset of NAFLD is hallmarked by the accumulation of lipids within hepatocytes (hepatic steatosis), which arises from an imbalance between triglyceride import, production and extrusion primarily caused by obesity and a hypercaloric diet. Furthermore, steatosis can be induced by a variety of drugs. Steatosis can progress into non-alcoholic steatohepatitis (NASH), an inflammatory condition that can result in liver failure. An additional and important cause of liver injury is adverse drug reactions (ADRs). It is estimated that ADRs cost as much as drug treatment itself, that it causes withdrawal of 4 % of new chemical entities from the market and is the 4th- 5th leading cause of death (cf. Eichelbaum et al., 2006). It has been estimated that adverse drug reactions cause up to 7 % of all hospital admissions in the UK and 13% of all admissions to internal medicine clinics in Sweden are due to ADRs (Cf. Sim and Ingelman-Sundberg, 2011).
Our research aims at understanding the basis for these liver diseases, drug induced injuries and the differences between individuals in liver function, risk for liver diseases, liver regeneration, human drug metabolism, drug toxicity and drug response, thereby facilitating a more effective personalized drug treatment at the clinics. The research encompasses:
- Identification and functional validation of genetic polymorphism of the genes encoding drug transporters, drug metabolizing enzymes and drug targets.
- Implementation of pharmacogenetics into psychiatry by the identification of novel genetic variants of importance for metabolism of CNS drugs and validation in large clinical cohorts.
- Identification of novel mechanisms for drug induced liver injury using 3D spheroid models.
- Identification of the role of different types of liver cells, signal transduction systems and mediators for development of NASH using 3D liver spheroids.
Projects
Mimicking liver function and liver disease in vitro for drug development
1.1 Summary
We use 3D hepatic spheroid systems obtained from human liver donations or cryopreserved hepatocyte cultures in combination with different types of non-parenchymal cells to:
- Evaluate the usefulness of these spheroid systems for studying properties and functions of diseased liver models and their potential for use during drug development including validation of existing drugs and development of new drugs.
- Evaluate in detail mechanisms of formations and treatment of NAFLD, NASH and fibrosis in the spheroids systems including screening for novel drug therapies using the spheroids as validation system.
- Evaluate chronic drug toxicity in diseased liver systems as well as novel mechanisms for drug induced hepatocyte proliferation and cytoskeletal changes.
This information will be of importance for:
- Increased possibilities to avoid the development of hepatotoxic drugs.
- Understanding different liver diseases and
- Validation of drug targets and drug candidates aimed for treatment of liver diseases.
1.2 Experimental platform
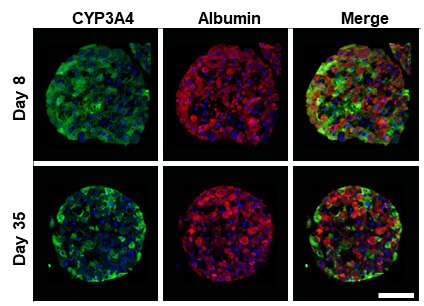
In our lab we have established and further developed novel spheroid based 3D systems for studies of liver function and properties in vitro (Gunness et al., 2013; Bell CC et al., 2016).
Starting with hepatocytes from commercial sources, we form spheroids from primary human hepatocyte fractions (PHH spheroids) using the ultra-low attachment plates and defined media. In the spheroids, the cells have relevant cell-cell interactions, a physiological cell-extracellular matrix (ECM) interface, improved cell polarity and a functional bile canaliculi network (see Figure 1). Indeed we showed that relevant enzyme expression can be preserved for at least 5 weeks in culture and that periportal and perivenous hepatocytes retain their phenotype during this period (Bell et al., 2016). The hepatocytes in 3D spheroids exhibit molecular phenotypes on transcriptomic, proteomic and metabolomic level similar to the livers from which they originate. We can transfect the spheroids with virus mimicking viral induced hepatitis or supplement the media with bile acids, allowing the faithful identification of drugs with cholestatic liabilities (Hendriks et al., 2016).
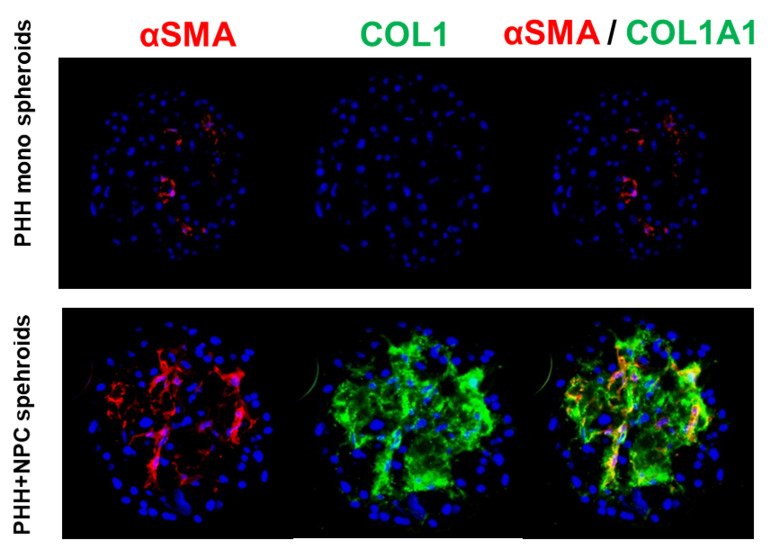
Moreover, we can induce steatosis by drugs or by altering the media composition and we can grow the spheroids in the presence of non-parenchymal cells and induce inflammation, hepatitis and liver fibrosis (Hurrell et al., 2020). We have also developed a model for detection of cholestatic drugs.
1.3 Pathological liver systems
Using these diseased liver systems we elucidate the drug hepatotoxicity including their mechanisms (Hendriks et al., 2019) in diseased liver as compared to healthy liver. We also evaluate novel mechanisms for drug mediated enzyme induction (Hendriks et al., 2020), study factors causing steatosis, and fibrosis in the liver and new regimens to inhibit such development.
The transition from steatosis to NASH, fibrosis and HCC is characterized by an intricate interplay of a multitude of nutritional and genetic factors, pathways and cell types. We validate the utility of the 3D spheroid culture system in which PHH can be co-cultured with Kupffer and stellate cells in 96-well format (Bell et al., 2016; Hurrell et al., 2020) as an in vitro disease model for human NAFLD compatible with high-throughput analyses.
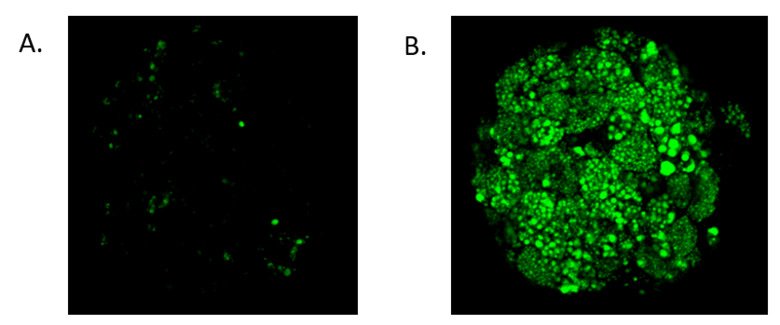
We develop culture conditions that closely mimic the nutritional and hormonal status of hepatocytes in vivo with regards to e.g. glucose and glucagon levels, fatty acids and insulin. In this system, we extensively characterize 3D cultured hepatocytes with regards to expression levels and activities of gene products involved in liver homeostasis and causing insulin resistance and metabolic diseases. We have previously shown that our human 3D culture system can recapitulate lipid accumulation, when exposed to elevated levels of fatty acids, fructose and insulin, as is the case in NAFLD in vivo (Figure 2).
Currently we strive hard to develop different models for liver fibrosis and evaluate drug candidates for their treatment.
1.4 Mechanisms for drug induced enzyme induction
In vivo in man drugs can cause induction of enzymes participating in their metabolism. Such induction can lead to resistance for drug treatment and it is important to evaluate preclinically that drug candidates do not harbour such a property. On the basis of a pharmacokinetic failure in clinical trials where animal models and conventional 2D hepatocyte models were unable to predict drug induced induction in vivo, we are, based on the 3D spheroid model, investigating novel mechanisms for drug dependent CYP induction which involves intracellular signal transduction systems. The 3D spheroid system is validated as a new standard in vitro system for prediction of drug mediated enzyme induction.
2. Role of CYP2C19 in brain phenotypes
The CYP2C19 enzyme metabolizes endogenous and synthetic psychoactive compounds including steroid hormones, cannabinoids, SSRIs, tricyclic antidepressants, and benzodiazepines. Two major variant alleles are known, CYP2C19*2 is defective and CYP2C19*17, which is identified by our group, is causing increased enzyme expression.
This project aims to reveal the role of CYP2C19 enzyme in prenatal brain development, as well as its role in adulthood in affective disorders and metabolism of psychopharmaceuticals.
2.1. Involvement of endogenous CYP2C19 in depressive phenotypes and hippocampal homeostasis
We detected CYP2C19 expression in fetal human brain and in collaboration with Nancy Pedersen, we previously showed that the presence of two defective (CYP2C19*2) alleles causes a decrease in depressive symptoms. We found that transgenic mice containing human CYP2C19 gene also show expression of this gene in developing brain, as well as decreased hippocampal volume, anxious phenotype, and increased hippocampal activation after acute stress. In humans, we found that the absence of CYP2C19 is associated with a bilateral hippocampal volume increase in two independent healthy cohorts and a lower prevalence of major depressive disorder and depression severity in African-Americans (N=3,848) (Jukic et al., 2016, 2017). Transgenic 2C19 mice showed high stress sensitivity, impaired hippocampal Bdnf homeostasis in stress, and more despair-like behavior in the forced swim test (FST). So indeed several of the phenotypes originally described to be caused by overexpression of CYP2C19 in mice were also found to be influenced by CYP2C19 in humans which indicates that elevated CYP2C19 expression is associated with depressive symptoms, reduced hippocampal volume and impairment of hippocampal serotonin and BDNF homeostasis.
2.2. Establishing of CYP2C19 genotype-driven dosing regimen of escitalopram
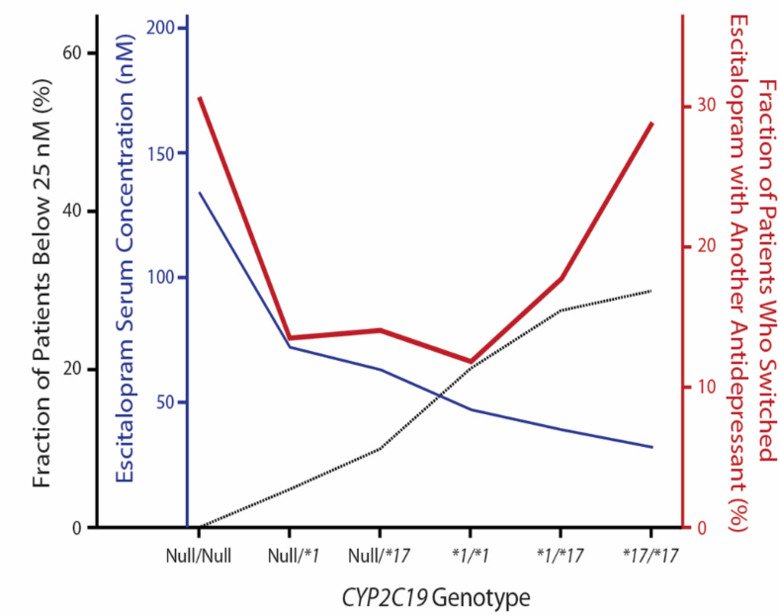
Well replicated in vitro and clinical studies indicate that escitalopram (ESC) is the most selective and most efficient among selective serotonin reuptake inhibitors (SSRIs). SSRIs are the cornerstone of the modern antidepressant pharmacotherapy; however, a large proportion of depressed patients do not respond adequately to the treatment with these drugs. The variation in the disposition of ESC due to CYP2C19 polymorphism may significantly contribute to the inter-individual variability in antidepressant response among the patients. In this project, we found a large contribution of CYP2C19 polymorphism to the escitalopram levels as well as the treatment success based on successful drug utilization as measured by the extent of drug switching (Fig 3).
2.3. Involvement of CYP2C19 in developing brain in physiology and degradation of dopaminergic neurons
Movement disorders, including Parkinson’s disease, significantly contribute to the morbidity caused by diseases worldwide; however their etiology and pathophysiology are still largely unclear. Animals overexpressing the human CYP2C19 gene (2C19TG) exhibit an increased dopamine (DA) concentration in the brain and hyperkinesia in the young age and degeneration of dopaminergic neurons in the old age. Conversely, healthy human subjects of genotype connected with elevated CYP2C19 expression show lower grey matter scores in substantia nigra (SN), indicative of DA neuronal degradation in this brain region. The aims of this project are to: (1) Track the state and function of DA neurons in SN of 2C19TG mice to understand the succession of their early hyperactivity and late (potential) degradation in a systematic manner, (2) Elucidate the alteration in biochemical cascades behind the reduction in dopaminergic neurons in 2C19TG mutants, and (3) Validate the clinical significance of these findings by using neuroimaging techniques on healthy individuals. Overall, the investigation through which CYP2C19 affects dopaminergic neuronal physiology and degradation is aimed to lead to the establishment of genetic and neuroimaging biomarkers, as well as to provide novel drug-targets for the treatment of dopamine-mediated motoric deficits.
3. Evaluation of the importance of rare genetic variants and novel haplotypes on hepatic metabolism and drug response
Genetic variants primarily encoding drug and metabolite transporters, phase I and phase II drug metabolizing enzymes and nuclear receptors can influence drug response by modulating drug absorption, distribution, metabolism and excretion (ADME). Importantly, while in the past decades an ever-growing arsenal of genetic variants with demonstrated impacts on human drug response has been identified in these pharmacogenes, a substantial fraction of the heritable variability in drug response remains unexplained. Rare genetic variants that only occur in very few individuals and are hence missed in genome-wide association studies have been proposed to contribute to this missing heritability. We have recently highlighted the role of CYP2D6 and CYP2C19 polymorphism for better treatment of schizophrenia and depression, respectively. The results indicate important effects on the clinical outcome by personally designing the drug dose based on the genetic variants of these enzymes the patients carry (Jukic et al, 2019; Jukic et al. 2018; Jukic et al., 2021; Milosavljevic et al., 2021).
In collaboration with Espen Moldens group in Oslo and Lili Milanis group at the Estonian Genome Center, Tartu, we currently identify novel genetic loci of high importance for personalization of drug treatment using large patient cohorts. The polymorphisms identified are evaluated in our hepatic spheroid system using gene silencing by e.g. siRNA constructs. We have recently found novel genetic loci for determining the metabolism of antidepressants and antipsychotics as well as novel haplotypes of CYP genes determining drug metabolism (Bråten et al., 2021). These results will allow the construction of novel microarrays for personalized drug treatment in the clinic.
This project is linked to our participation in the H2020 project Ubiquitous pharmacogenomics (U-PGx; http://upgx.eu/) in which we have the primary responsibility to analyze the genetic background of outlier patients with abnormal drug response.
Publications
Selected publications
- Article: BRITISH JOURNAL OF CLINICAL PHARMACOLOGY. 2024;90(3):740-747Wollmann BM; Smith RL; Kringen MK; Ingelman-Sundberg M; Molden E; Storset E
- Article: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2023;114(3):673-685Kloditz K; Tewolde E; Nordling A; Ingelman-Sundberg M
- Article: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2023;113(6):1284-1294Jarvinen E; Hammer HS; Poetz O; Ingelman-Sundberg M; Stage TB
- Article: NEUROPATHOLOGY AND APPLIED NEUROBIOLOGY. 2023;49(1):e12867Milosavljevic F; Brusini I; Atanasov A; Manojlovic M; Vucic M; Orescanin-Dusic Z; Brkljacic J; Miljevic C; Nikolic-Kokic A; Blagojevic D; Wang C; Damberg P; Pesic V; Tyndale RF; Ingelman-Sundberg M; Jukic MM
- Article: CELLS. 2023;12(2):302Trampuz SR; van Riet S; Nordling A; Ingelman-Sundberg M
- Article: CTS-CLINICAL AND TRANSLATIONAL SCIENCE. 2023;16(1):62-72Lenk HC; Lovsletten Smith R; O'Connell KS; Jukic MM; Kringen MK; Andreassen OA; Ingelman-Sundberg M; Molden E
- Review: TRENDS IN PHARMACOLOGICAL SCIENCES. 2022;43(12):1055-1069Jukic M; Milosavljevi F; Molden E; Ingelman-Sundberg M
- Editorial: TRENDS IN PHARMACOLOGICAL SCIENCES. 2022;43(12):987-993Ingelman-Sundberg M; Cadet JL; Gobbi G; Olson DE; Yohn S; Foster DJ
- Article: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2022;112(5):1000-1003Pridgeon CS; Johansson I; Ingelman-Sundberg M
- Article: CTS-CLINICAL AND TRANSLATIONAL SCIENCE. 2022;15(9):2135-2145Braten LS; Ingelman-Sundberg M; Jukic MM; Molden E; Kringen MK
- Article: PHARMACOGENOMICS JOURNAL. 2022;22(4):247-249Haeggstrom S; Ingelman-Sundberg M; Paabo S; Zeberg H
- Article: CELLS. 2022;11(10):1597Pridgeon CS; Bolhuis DP; Milosavljevic F; Manojlovic M; Vegvari A; Gaetani M; Jukic MM; Ingelman-Sundberg M
- Article: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2022;111(5):1165-1174Lenk HC; Kloditz K; Johansson I; Smith RL; Jukic M; Molden E; Ingelman-Sundberg M
- Article: ADVANCES IN PHARMACOLOGY. 2022;95:393-416Ingelman-Sundberg M
- Article: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2021;110(3):750-758Jukic MM; Smith RL; Molden E; Ingelman-Sundberg M
- Article: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2021;110(3):786-793Braten LS; Haslemo T; Jukic MM; Ivanov M; Ingelman-Sundberg M; Molden E; Kringen MK
- Review: INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. 2021;22(15):8221Harjumaki R; Pridgeon CS; Ingelman-Sundberg M
- Article: SCIENCE TRANSLATIONAL MEDICINE. 2021;13(603):eabf3637van der Lee M; Allard WG; Vossen RHAM; Baak-Pablo RF; Menafra R; Deiman BALM; Deenen MJ; Neven P; Johansson I; Gastaldello S; Ingelman-Sundberg M; Guchelaar H-J; Swen JJ; Anvar SY
- Article: DRUG METABOLISM AND DISPOSITION. 2021;49(7):DMD-AR-2020-000340-508Riede J; Wollmann BM; Molden E; Ingelman-Sundberg M
- Review: JAMA PSYCHIATRY. 2021;78(3):270-280Milosavljevic F; Bukvic N; Pavlovic Z; Miljevic C; Pesic V; Molden E; Ingelman-Sundberg M; Leucht S; Jukic MM
- Article: SCHIZOPHRENIA RESEARCH. 2021;228:590-596Smith RL; Tveito M; Kylleso L; Jukic MM; Ingelman-Sundberg M; Andreassen OA; Molden E
- Review: ARCHIVES OF TOXICOLOGY. 2021;95(1):11-25Smutny T; Hyrsova L; Braeuning A; Ingelman-Sundberg M; Pavek P
- Article: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2020;108(4):844-855Hendriks DFG; Vorrink SU; Smutny T; Sim SC; Nordling A; Ullah S; Kumondai M; Jones BC; Johansson I; Andersson TB; Lauschke VM; Ingelman-Sundberg M
- Article: ADVANCED SCIENCE. 2020;7(15):2000248Oliva-Vilarnau N; Vorrink SU; Ingelman-Sundberg M; Lauschke VM
- Article: CELLS. 2020;9(4):E964-964Hurrell T; Kastrinou-Lampou V; Fardellas A; Hendriks DFG; Nordling A; Johansson I; Baze A; Parmentier C; Richert L; Ingelman-Sundberg M
- Review: PHARMACOLOGY & THERAPEUTICS. 2019;197:122-152Lauschke VM; Zhou Y; Ingelman-Sundberg M
- Article: LANCET PSYCHIATRY. 2019;6(5):418-426Jukic MM; Smith RL; Haslemo T; Molden E; Ingelman-Sundberg M
- Article: PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA. 2019;116(1):217-226Cordero-Herrera I; Kozyra M; Zhuge Z; Haworth SM; Moretti C; Peleli M; Caldeira-Dias M; Jahandideh A; Han H; Cruz JDC; Kleschyov AL; Montenegro MF; Ingelman-Sundberg M; Weitzberg E; Lundberg JO; Carlstrom M
- Article: SCIENTIFIC REPORTS. 2018;8(1):14297Kozyra M; Johansson I; Nordling A; Ullah S; Lauschke VM; Ingelman-Sundberg M
- Article: GENETICS IN MEDICINE. 2018;20(6):622-629Santos M; Niemi M; Hiratsuka M; Kumondai M; Ingelman-Sundberg M; Lauschke VM; Rodriguez-Antona C
- Article: HUMAN GENOMICS. 2018;12(1):26Ingelman-Sundberg M; Mkrtchian S; Zhou Y; Lauschke VM
- Article: AMERICAN JOURNAL OF PSYCHIATRY. 2018;175(5):463-470Jukic MM; Haslemo T; Molden E; Ingelman-Sundberg M
- Review: ANNUAL REVIEW OF PHARMACOLOGY AND TOXICOLOGY. 2018;58:161-185Lauschke VM; Barragan I; Ingelman-Sundberg M
- Article: MOLECULAR PSYCHIATRY. 2017;22(8):1155-1163Jukic MM; Opel N; Stroem J; Carrillo-Roa T; Miksys S; Novalen M; Renblom A; Sim SC; Penas-Lledo EM; Courtet P; Llerena A; Baune BT; de Quervain DJ; Papassotiropoulos A; Tyndale RF; Binder EB; Dannlowski U; Ingelman-Sundberg M
- Article: DRUG METABOLISM AND DISPOSITION. 2017;45(4):419-429Bell CC; Lauschke VM; Vorrink SU; Palmgren H; Duffin R; Andersson TB; Ingelman-Sundberg M
- Article: HEPATOLOGY. 2016;64(5):1743-1756Lauschke VM; Vorrink SU; Moro SML; Rezayee F; Nordling A; Hendriks DFG; Bell CC; Sison-Young R; Park BK; Goldring CE; Ellis E; Johansson I; Mkrtchian S; Andersson TB; Ingelman-Sundberg M
- Article: SCIENTIFIC REPORTS. 2016;6:35434Hendriks DFG; Puigvert LF; Messner S; Mortiz W; Ingelman-Sundberg M
- Article: NUCLEIC ACIDS RESEARCH. 2016;44(14):6756-6769Ivanov M; Kals M; Lauschke V; Barragan I; Ewels P; Kaller M; Axelsson T; Lehtio J; Milani L; Ingelman-Sundberg M
- Article: SCIENTIFIC REPORTS. 2016;6:25187Bell CC; Hendriks DFG; Moro SML; Ellis E; Walsh J; Renblom A; Puigvert LF; Dankers ACA; Jacobs F; Snoeys J; Sison-Young RL; Jenkins RE; Nordling A; Mkrtchian S; Park BK; Kitteringham NR; Goldring CEP; Lauschke VM; Ingelman-Sundberg M
- Show more
Full list of publications
Full list of publications from Magnus Ingelman-Sundberg.
Funding
- Swedish Research Council
- European Union, H2020 and FP7 (U-PGx)
- Department of Physiology and Pharmacology/Karolinska Institutet
- The Swedish Brain Foundation (Hjärnfonden)
- NovoNordisk
- ERC (Advanced grant to MIS)
Staff and contact
Group leader
- Magnus Ingelman-SundbergProfessor, Senior
All members of the group
- Andrea AtanasovAffiliated to Research
- Inger JohanssonSenior Lecturer
- Marin JukicSenior Research Specialist
- Åsa NordlingLaboratory Assistent
- Patrick RiekerAffiliated to Research
- Aileen TranAffiliated to Research
- Arnau Vila BertomeuAffiliated to Research
- Sander van RietPostdoctoral Researcher
Visiting address
Karolinska Institutet, Biomedicum, Quarter B5, Solnavägen 9, Stockholm, 171 65, Sweden
Postal address
Karolinska Institutet, Department of Physiology and Pharmacology, Ingelman-Sundberg group - Pharmacogenetics, att: (name of recipient), Stockholm, 171 71, Sweden