Our research
Epigenetics is a rapidly expanding research field ranging from basic research to clinical applications. The term epigenetics comes from developmental biology and is a theory developed by Conrad Waddington in the 1940s to explain how genes are used during the process of morphogenesis and development of a multicellular organism (the process of epigenesis). Since all somatic cells of an organism, with very few exceptions, contain exactly the same DNA sequence, epigenetics can possibly explain how the same DNA sequence can be used to express different genes in different cell types and tissues. A more modern definition of epigenetics is ‘nuclear inheritance’ which is not based on differences in the DNA sequence. With this we now consider all modifications of the chromatin template that together establish and propagate different stable patterns of gene expression.
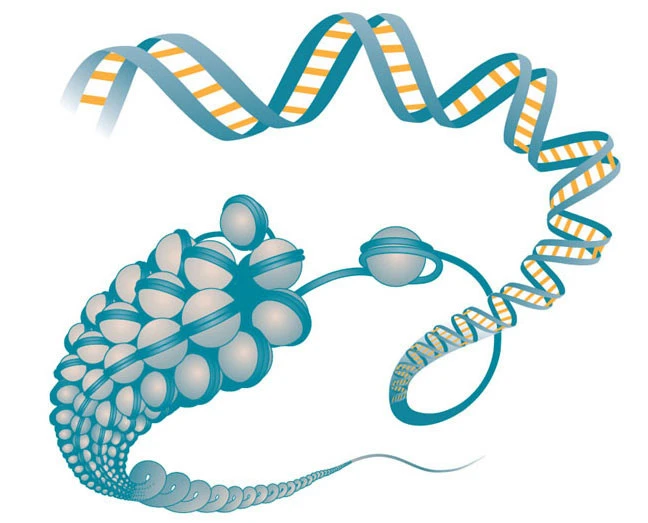
Epigenetic modifications
Covalent chemical modifications to the DNA and to histones, histone variants, nucleosome positions, non-coding RNAs and the level of chromatin compaction all contribute to chromosomal structure and to the activity or silencing of genes. These chromatin-level alterations are defined as epigenetic when they are heritable from mother to daughter cell. Histone acetylation and methylation are the most common modifications and they occur at specific lysines mainly in the NTER region of histones H3 and H4. The modifications lead to functional changes in gene promoters and enhancers because they confer altered accessibility to transcriptional regulatory complexes, thus impacting on gene expression. Nucleosome remodelling activity of SNF2 enzymes results in nucleosome sliding, modulation of histone turnover, spacing of nucleosomes, histone eviction and histone exchange. These activities are central in epigenetic regulation and control accessibility to the DNA for a large number of factors involved in processes involving recognition of the DNA sequence i.e. RNA transcription, DNA replication, repair and recombination.
The powerful molecular genetic analysis in fission yeast (Schizosaccharomyces pombe) is important for dissecting the detailed molecular function of several epigenetic mechanisms including histone acetylation, histone methylation and SNF2 chromatin remodelling. Our work in human cell lines allows for identification of conserved epigenetic mechanisms and the clinical epigenetics projects show a relevance of epigenetic mechanisms in disease.
