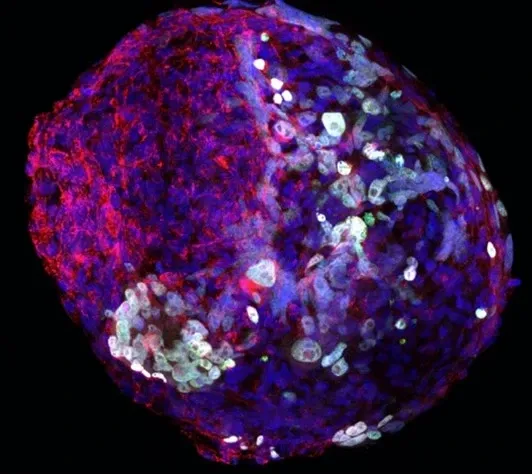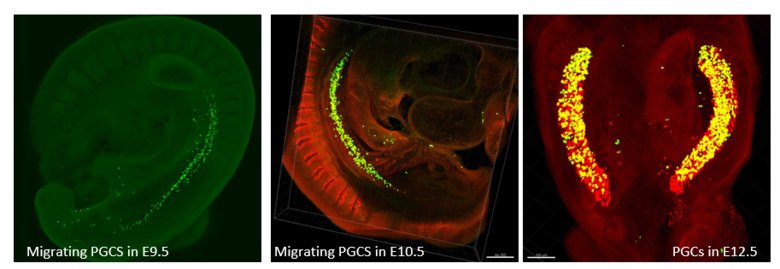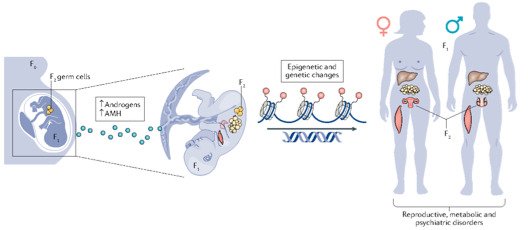
Our research group is interested in the developmental principles of germline specification in health and disease using mouse models and human stem cell cultures coupled with state-of-the-art molecular and cellular tools. Moreover, we investigate how maternal diseases conditions impact the health outcomes of future offspring through the process called developmental programming by placentas and/or germline modulation, a process known as epigenetic inheritance of disease or developmental origins of health and disease (DOHaD). Our current research focuses on polycystic ovary syndrome (PCOS) and diabetes in women taking advantage of disease mouse models, human cohort samples, single-cell sequencing and disease-derived placental organoids in microfluidic co-culture system.
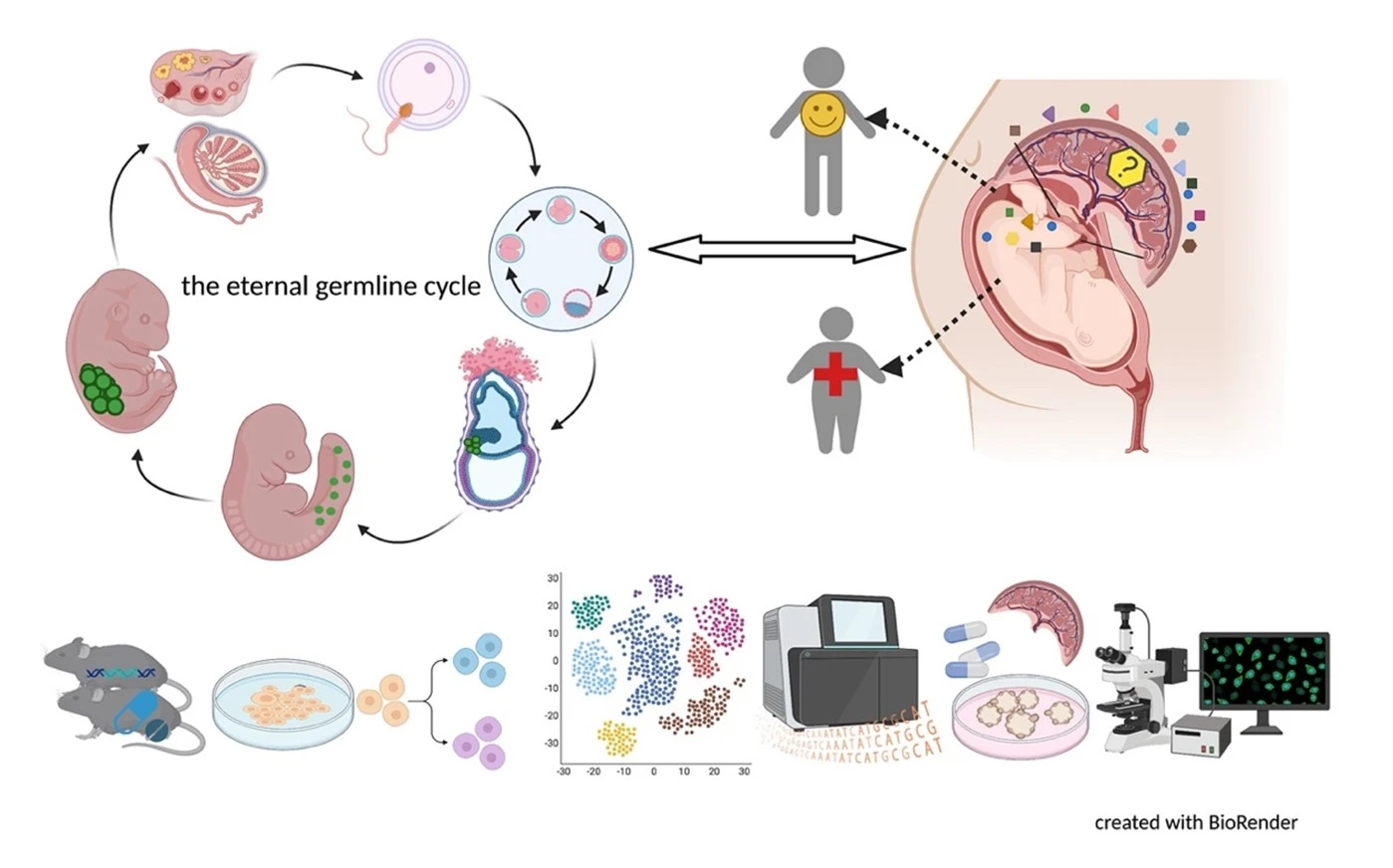
Germ cells are often considered to be immortal, as they serve as the sole carriers of genetic and epigenetic information across generations, perpetuating life. The germline cycle is a lengthy process, beginning with early gastrulation and continuing through gametogenesis after birth. Any errors that occur during this process can have devastating and long-lasting effects. Recent advances in single-cell sequencing technology have greatly expanded our knowledge of germline development in mammals, but many questions remain unanswered. In this article, we aim to address some of these questions, such as (1) what regulates progenitor competence and how can we define cell quality for germline specification? (2) How is germline specification correlated with epigenetic remodeling including X-chromosome dosage effects? (3) How do certain genetic mutations affect gametogenesis?
Interestingly, there are two waves of epigenetic remodeling that occur after fertilization. These processes ensure the totipotency of the blastocyst for somatic lineage specification and establish a "clean slate" for germ cells, erasing any potentially harmful epigenetic modifications acquired from parents. These processes allow the organism to adapt to changing environmental conditions and minimize the risk of inheriting harmful traits. However, the completeness and faithfulness of these processes still need to be investigated. Increasing evidence shows that parental health conditions can predispose their offspring to develop diseases such as obesity, diabetes, cardiovascular disease, and behavioral disorders through developmental programming, a process called epigenetic inheritance of diseases. Mechanistic understanding of these processes is still sparse. We aim to answer questions such as (1) how do parental health conditions affect the germline, which further transmits phenotypic traits? (2) how does the placenta respond to adverse uterine environments, which can systematically modify the cellular and physiological functions of the developing fetues? (3) can we systematically model maternal disease signatures with offspring key organ signatures in humans using organoids and microfluidic coculture?
We are among those pioneers to apply and develop single-cell RNA sequencing (Smart-seq, Smart-seq2, LCM-seq etc). More tools to answer all these interesting questions are mouse disease models, human iPSC culture and differentiation, organoid culture, human sample cohorts and registry data together with other key cellular and molecular assays.
News
 Photo: Creative Commons CC0
Photo: Creative Commons CC0Make a donation to our research
Your support means a lot to the success of research. This allows us to go further in our efforts to improve human health through research and education.