
Our research
A prerequisite for maintained genome integrity during cell divisions depends on correct gene expression, DNA replication, DNA repair as well as chromosome structure and segregation. The members of the family of conserved Structural Maintenance of Chromosome (SMC-) complexes have overlapping and unique functions during all these processes. Thus, the SMC complex Cohesin, in focus of this project, is necessary for cohesion between sister chromatids until anaphase, thereby promoting high-fidelity chromosome segregation. In addition, it is essential for repair of various types of DNA damage, as well as for shaping the genome in 3D. Deficiency at any level of the Cohesin pathway frequently leads to aneuploidy and genome instability, tumorigenesis and cancer. Indeed, the last ten years it has become clear that Cohesin proteins are among the most frequently mutated in Cancer.
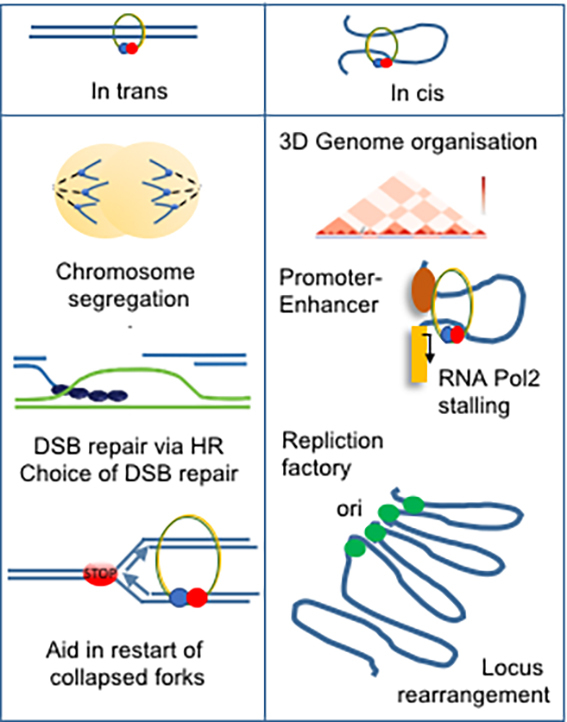
In addition, a group of developmental syndromes called Cohesinopathies are caused by mutations in several of the Cohesin subunits or its regulators. Clinical isolates from these patients are also found to be highly DNA damage sensitive. This together motivates our studies trying to understand the mechanism for tumour development, and Cohesinopathies, driven by deficient DNA damage response and malfunctions of the Cohesin network. Using yeast and human cells as model systems, combined with advanced biochemistry, functional studies and genomic approaches we expect that our studies will open up a new era for understanding the molecular mechanisms for Cohesin during both healthy and malignant cell cycles as well as DNA repair events, and provide novel treatment strategies for Cancer as well as support to CdLS patients. To progress towards these goals, we work on:
- Understanding the molecular mechanisms for the role of the Cohesin network during DSB response and repair, in connection to local and global chromatin structure, as well as transcription
- Determining why and how DSB induction activates de novo cohesion establishment, and if this Damage-Induced cohesion is essential for telomere maintenance
- Evaluating if manipulation of the Cohesin network is a possible new target for Cancer treatment
