Our research
Acute myeloid leukaemia (AML) has a poor prognosis in both, adults and children. Acute leukemia accounts for 30% of the diagnosed cases of pediatric cancer. Acute lymphoblastic leukemia (ALL) is the most common subtype in children and has good prognosis with 80-90% survival. However, pediatric and infant acute myeloid leukemia still have a poor outcome. There is therefore an urgent need for new novel therapeutics. Dramatic improvements in treatment and outcome have been made, when the biology is well-characterised. For acute promyelocytic leukaemia (APML) the long-term survival now exceeds 90%, demonstrating that a better understanding of AML biology is a pre-requisite for the development of novel therapies to improve treatment outcomes. AML is characterised by early mutations and chromosomal aberrations in epigenetic regulators and transcription factors. Therefore, we are focusing our research on epigenetic regulation and epigenetic treatment of AML in adults children and infants.
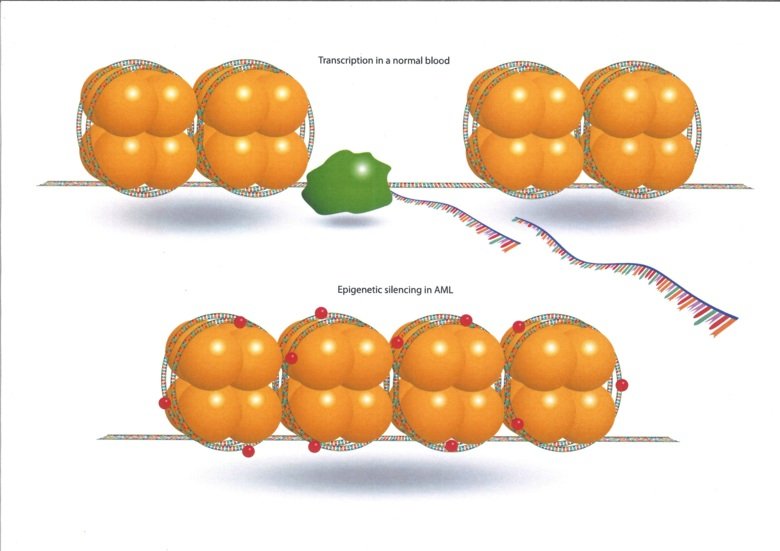
We are using cutting edge genome-wide technologies, biochemistry and molecular biology methods, novel model-systems and primary patient material, to obtain an increased knowledge of the molecular mechanisms that drive AML. The results are used to identify new drugs and drug combinations to develop improved treatments for AML. We are working together with several international research groups who are developing technologies to be able to analyse transcriptomics and epigenomics in new ways and in small cell populations, such as the leukemic stem cells. In addition, we have close collaborations with clinical haematologists to be able to ask the clinically important research questions. Within this network of technical know how, clinical expertise and scientific drive, we have the tools to make major advances in the treatment of AML.
Dysregulated metabolic enzymes mediate vulnerabilities for disease via epigenetic mechanisms
The molecular interactions and crosstalk between epigenetic regulation and cellular metabolism are significantly influenced by diet and metabolism and thus are dependent on nutrient availability. Recently an intersecting mechanism has been described, suggesting that there are under-studied molecular mechanisms that facilitate epigenomic dynamics under perturbed metabolic conditions that may contribute to human disease.
Our projects include:
- Epigenetic regulations of normal blood development, to better understand the perturbations found in AML.
- Identifying epigenetic disturbance in AML patients.
- Using iPS cells retrieved from AML patients as a model system to understand the molecular development of AML.
- Testing and analysing the drug response of novel drug or drug combinations, both in vitro and by participating in clinical trials.
- Dissect the metabolic-epigenetic molecular networks in blood cells; highlight their role in mediating cellular responses; and explore their modifiability through dietary and pharmacological interventions for metabolic associated diseases.
Work environment and values
Podcast: Creating Psychological Safety in the Workplace from HBR IdeaCast.
Developed Tools and databases
EpiFactors - a tool for cell specific expression of epigenetic regulators.

Looking for a Postdoc or Master position?
We are always interested to recruit motivated students and postdocs!
There is currently a PhD position announced through the MSCA Doctoral Network ACHILLES.
