Research at Helin Norberg lab
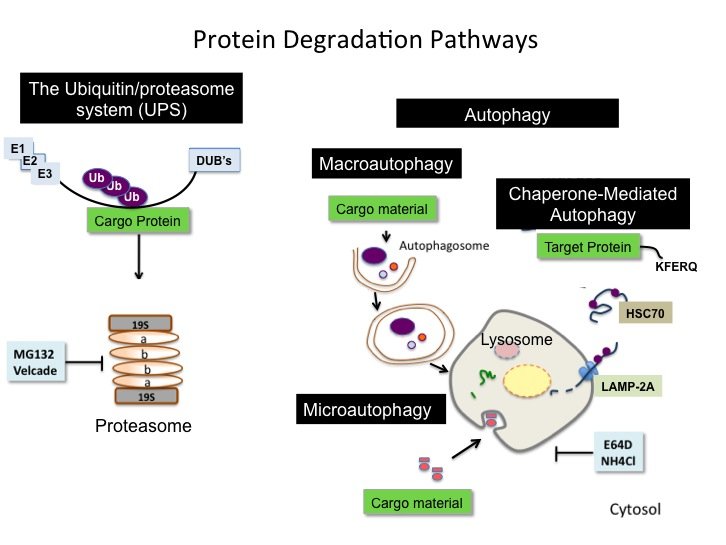
The Norberg lab current research focuses on Chaperone-mediated Autophagy (CMA), a unique type of mammalian autophagy that only applies to proteins without affecting cellular organelles.
Over the recent years, the Norberg lab has explored new therapeutic approaches by stimulating the cell’s own quality control mechanisms to induce the degradation of pathogenic proteins to prevent their aberrant accumulation. More recently, the lab has been dedicated on developing a new field of `Controlled Autophagic Proteostasis' by studying the autophagy-lysosomal pathway. Therefore, the current research focuses on Chaperone-mediated Autophagy (CMA), a unique type of mammalian autophagy that only applies to proteins without affecting cellular organelles.
The role and impact of CMA in Cancer
Proteins that promote tumor growth and dissemination are often mutated, deregulated and distinctively stabilized in cancer cells. As pathogenic accumulation of these proteins effectively sustains cancer progression, they represent indisputable targets for treatment. Hence, understanding how these proteins can be removed is of great importance for improved cancer therapy. The goal of this project is therefore, to study mechanisms of protein degradation by CMA to understand a) the implication of recruiting the unique type of mammalian autophagy to specifically promote lysosomal degradation of oncoproteins, b) to identify novel CMA targetable oncoproteins, c) the impact of degrading oncoproteins by CMA.
To explore the impact of CMA activation
The role of CMA in cancer cells remain paradoxal and the physiological importance of CMA in cancer is currently not defined. Therefore, this project intends to investigate the CMA pathway in depths both at cellular and organismal level, in order to explore the intriguing possible role of activating CMA in various human malignancies to find selective targeted approaches applicable for pathogenic proteins. Further the project aims to understand the underlying molecular mechanisms of CMA activation and its oncogenic and pathogenic targeting. Ultimately, we aim to explore the impact of CMA activation in vivo.
The role and impact of CMA in human disorders
The findings from the above-mentioned projects will provided with answers to address a long-standing question in cell biology; How is CMA activated and regulated in cells? However, although the answers to these questions will provide us with a detailed molecular understanding of a fundamental biological process, we still need a comprehensive understanding of the role of CMA in various medical conditions as a basis for proposing new treatment strategies. Therefore, this project, is design to understand the role and impact of CMA in proteopathies and multiple other human diseases. This project has a translational focus which enables a tremendous potential to open up new therapeutic avenues and biomarkers.