Human life starts as a single cell that divides numerous times to create an entire human body consisting of trillions of cells. To maintain genome stability, cells need to faithfully replicate their DNA before dividing. A key feature of cancer cells is their forced proliferation, which causes a hasty uncontrolled start of DNA replication. To date, however, little is known about the mechanisms that control the start of DNA replication in human cells, hampering the design of better, selective cancer therapies. Studying essential, dynamic processes such as DNA replication requires conditional, time-resolved genetics. The Lemmens group develops and combines innovative protein depletion methods with cutting-edge imaging techniques to study how, when and where DNA replication starts. In parallel, we use various CRISPR-based technologies to discover new factors driving cell proliferation and cancer drug resistance.
Research projects
We develop innovative approaches to study DNA replication with high precision in both time and space. To determine the mechanism and kinetics of human DNA replication we run various projects, ranging from targeted imaging approaches to unbiased genetic screens.
We currently focus on three projects:
- To study dynamic processes in human cells at single-cell resolution, our team develops rapid protein depletion methods and time-resolved analysis pipelines.
- To define where these processes take place in the cell, we develop cutting-edge imaging approaches, including live-cell, high-content, super resolution and/or expansion microscopy.
- Many of our projects involve CRISPR technologies, including gene knockouts, reporter knock-ins and gene editing approaches. We also use pooled CRISPR screens to study how human cells become resistant to cancer drugs and discover new factors controlling genome maintenance or DNA replication.
Interested in joining our team? Please feel free to email Bennie Lemmens.
Collaborations
Understanding human biology and finding innovative cancer cures is a global team effort. We thus aim to collaborate with scientists around the world and currently join forces with i.a.:
- Dr. Thanos Halazonetis (University of Geneva, Switzerland)
- Dr. Hans Blom (Advanced Light microscopy Facility; KTH, Sweden)
- Dr. Mirek Dundr (Rosalind Franklin University, USA)
- Dr. Alexander van Oudenaarden (Hubrecht Institute, Netherlands)
- Dr. Bernard Schmierer (Genome Engineering Facility; KI, Sweden)
- Dr. Jiri Bartek (KI and Danish Cancer Society; Sweden and Denmark)
News
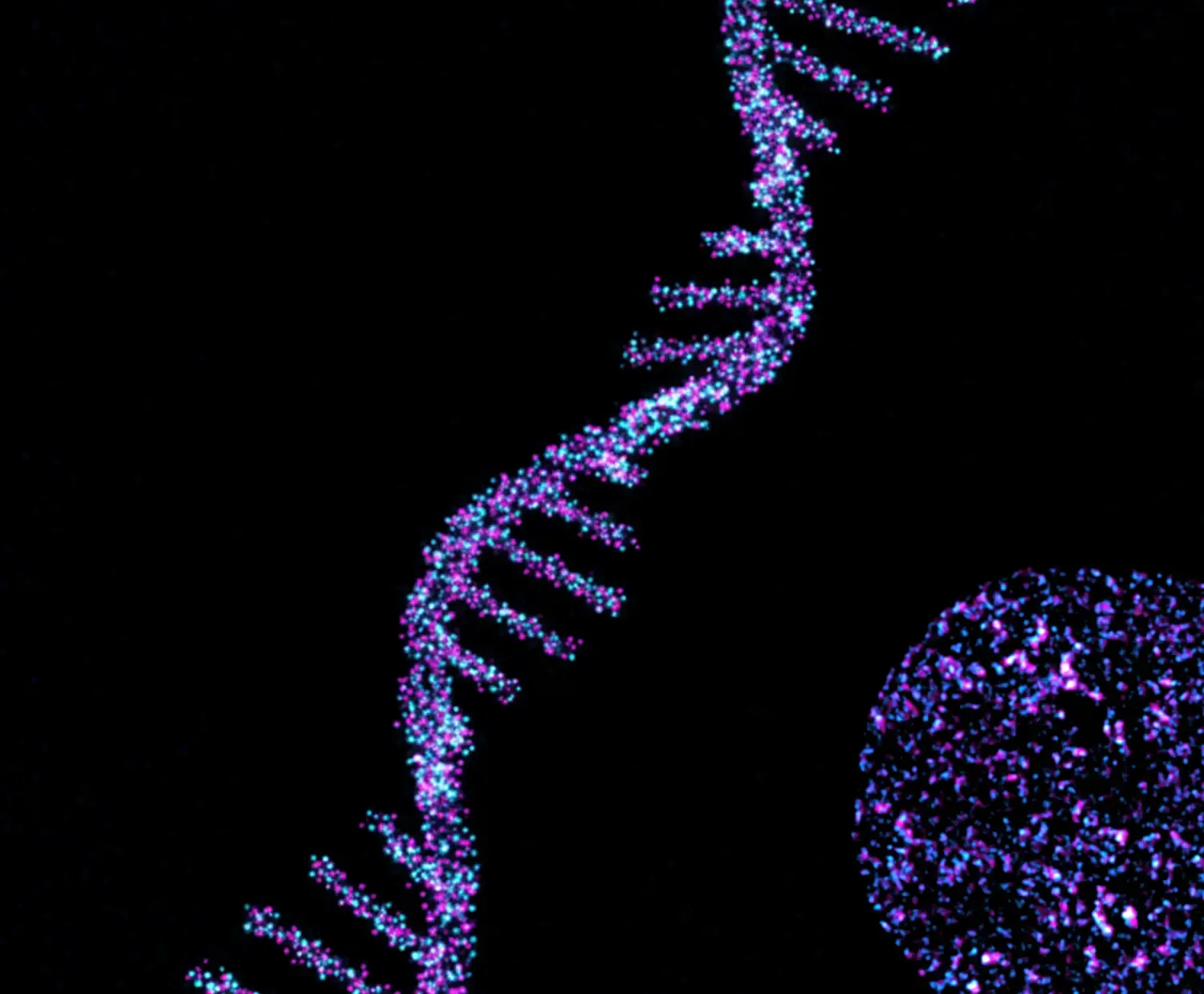 Photo: N/A
Photo: N/AASPIRE Awardees Uncover DNA Replication Dynamics with Unprecedented Precision
 Photo: N/A
Photo: N/A
