
- >
- >
- Amyotrophic Lateral Sclerosis - ALS – Caroline Ingre's research group
- Saving the brain in critically ill neonates – Ulrika Ådén's research group
- Regulatory Transcriptomics – Kutter group
- NeuroimAging – Grégoria Kalpouzos group
- Section of Pharmacogenetics – MIS-lab
- Biological and clinical aspects of the myelodysplastic syndrome – Magnus Tobiasson group
- Human innate lymphoid cell lab – Jenny Mjösberg group
- Viruses and their interactions with the immune system – Sara Gredmark Russ group
- Protein degradation pathways – Helin Norberg's research group
- Translational Auditory Neuroscience – Christopher Cederroth's research group
- Neuroradiology: Neurodegeneration and Neuroinflammation – Tobias Granberg's research group
- Tumor Immunology and Immunotherapy Group – Dhifaf Sarhan
- Causal inference in epidemiologic research – Arvid Sjölander's research group
- The role of autophagy in Aβ metabolism and neurodegeneration in Alzheimer’s disease – Per Nilsson group
- Darreh-Shori's Lab/group
- PROCOME – Hanna Öfverström's and Marta Roczniewska's group
- Molecular brain imaging of neurodegenerative disorders – Andrea Varrone's research group
- Alcohol and Drug Epidemiology – Mats Ramstedt's research group
- Anxiety related disoders and genetics – Christian Rück's research group
- Psychotherapy Research – Tobias Lundgren's research group
- Human olfaction – Mats J. Olsson's research group
- Tissue stem cells and aging – Pekka Katajisto's Group
- SOSVASC – David Lindström's research group
- Yenan Bryceson group
- Epidemiology of sarcoidosis and systemic lupus erythematosus – Team Elizabeth Arkema
- Viral genomics and metagenomics – Tobias Allander Group
- Defining the metabolic signatures of tissue resident human ILCs – Chris Tibbitt team
- Human papillomavirus which causes cervical cancer – Research Group Karin Sundström
- Research groups in bioinformatics and systems biology
- Cognitive epidemiology – Bo Melin's research group
- Gilad Silberberg group
- Research groups in oto-rhino-laryngology and logopaedics
- Immunoregulation of environmental-induced airway inflammation in severe asthma and bronchiectasis – Apostolos Bossios's research group
- Multimorbidity and frailty in older adults – Davide Liborio Vetrano group
- Psychiatric and pharmaco-epidemiology – Zheng Chang's research group
- Charting normal and malignant hematopoiesis – Team Joakim Dahlin
- Molecular mechanisms governing oxygen homeostasis in development, evolution, and disease – Kragesteen's research group
- Precision Medicine – Infrastructure team
- Research groups in medical epigenetics and epigenomics
- Spinal circuits for whole-body motor coordination – Laurence Picton group
- Clinical HIV epidemiology and comorbidity – Christina Carlander Research group
- Head and neck pathology – Anders Näsman's Team
- Carin Håkansta's research group – Digital work environment and health
- Pediatric and respiratory epidemiology – Catarina Almqvist Malmros' research group
- Sexual and Reproductive Health – Marie Klingberg-Allvin research group
- Angiogenesis, Cancer, Metabolic disease, Cardiovascular disease, Eye disease – Yihai Cao Group
- Cardiovascular and psychosocial health in resilience and healthy brain aging – Chengxuan Qiu Group
- Translational Psychiatry – Catharina Lavebratt's research group
- Rare metabolic liver diseases, coagulation and hepatocellular carcinoma – Stål and Wahlin research group
- Blood Engineering – Vanessa Lundin group
- Development, regulation and function of human innate lymphoid cells throughout life – Jakob Michaëlsson group
- Clinical Genetics – Richard Rosenquist Brandell's research group
- Oxysterols – Ingemar Björkhem and Ulf Diczfalusy research
- Molecular exercise physiology – Carl Johan Sundberg's research group
- Large-scale Network Connectivity in the Human Brain – Peter Fransson's research group
- Environmental physiology
- The Systems Virology Lab – Ujjwal Neogi
- Cancer Cell Invasion – Marco Gerling's research group
- Molecular brain imaging, neuropsychopharmacology, depression and anxiety disorders – Johan Lundberg's research group
- Organization and operation of postural neuronal networks in health and disease – Tatiana Deliagina group
- Paediatrics – Anna Lindholm Olinder's research group
- Petzold group
- Molecular and epidemiological studies of ascending aortic aneurysms – Research group Hanna Björck
- Prevention, Intervention and Mechanisms in Public Health (PRIME Health) – Cecilia Magnusson's research group
- The interplay between circadian 3D genome organization and metabolism in complex diseases – Anita Göndör's Group
- Reconstructive Plastic Surgery and Global Surgery – Jenny Löfgren's research group
- Real-world effectiveness of psychopharmacological treatment – Jari Tiihonen's Research Group
- Center for Heart Failure and Arrhythmia – Gianluigi Savarese
- Sports Medicine – Anders Stålman's research group
- Women's Mental Health Epidemiology – Donghao Lu Lab
- The SMC complexes, chromosome dynamics and stability – Camilla Björkegren's Group
- Extracellular vesicles in immunity – Research group Susanne Gabrielsson
- Precision Cancer Medicine in Lung Cancer - Preclinical, Translational & Clinical Research – Simon Ekman's research group
- RNA-guided DNA repair and cancer – Marianne Farnebo's research group
- Laboratory of translational fertility preservation – Kenny Rodriguez-Wallberg's group
- Research groups in cancer and oncology
- Common Mental Disorders and Behavioral Medicine in Primary Care – Erik Hedman's research group
- Neuronal membrane trafficking – Oleg Shupliakov group
- Research groups in paediatrics
- Biostatistics With Focus on Statistical Methods in Cancer Epidemiology and Screening – Keith Humphreys' research group
- Research groups in nanotechnology
- Gastroenterology – clinical epidemiology and clinical trials – Research group Ola Olén
- Precision engineering of T cells in vivo for therapies of cancer – William Nyberg team
- Protein Misfolding and Prevention by Molecular Chaperones in Cancer and Neurodegenerative Disease – Gefei Chen's research group
- Social Perception – Arvid Guterstam's research group
- Neuroepidemiology – Kyla McKay and Katarina Fink's research group
- Experimental growth research – Lars Sävendahl's research group
- Neuronal circuits of the Basal Ganglia – Maya Ketzef group
- Psychological Interventions - innovation, improvement and implementation – Viktor Kaldo's research group
- Susanna Ranta team
- Neurovascular diseases – Christina Sjöstrand research group
- Lifestyle, Prevention and Health – Ylva Trolle Lagerros research group
- Elias Arnér research group
- Gynecologic Cancer – Henrik Falconer's research group
- B cell biology, from immunodeficiency to cancer – Pan Hammarström Lab
- Developmental Cognitive Neuroscience – Torkel Klingberg group
- Rare Diseases – Ann Nordgren och Anna Lindstrand's research group
- Research on steatotic liver disease – Hagström Group
- Stem Cell Size – Jette Lengefeld Team
- Natural Killer Cell Biology and Cell Therapy – Kalle Malmberg group
- Mast Cell Biology – Research group Gunnar Nilsson
- Traumatic Brain Injuries and Neuromonitoring – Eric Thelin's research group
- Medical Statistics team
- Augmented Neurosurgery – Erik Edström and Adrian Elmi Terander's research group
- Molecular neurophysiology – Karima Chergui's research group
- Injuries Social Aetiology and Consequences (ISAC) – Lucie Laflamme's research group
- Unconventional T cells in human pathology – Andrea Ponzetta team
- Inflammation and immune regulation during infection – Team Christopher Sundling
- Orthopaedics – Anders Enocson's research group
- Endothelial dysfunction in atherosclerotic cardiovascular disease – Research Group John Pernow
- Clinical epidemiology of multiple sclerosis and inflammatory joint diseases – Team Thomas Frisell
- Cancer stem cells and clonal structure in acute lymphoblastic leukemia – Martin Enge's Group
- Mitochondrial metabolism in health and disease – Anna Wredenberg group
- Biomolecular Medicine and Advanced therapies- focus on delivery of RNA therapeutics – Research group Samir EL Andaloussi
- Antimicrobial resistance – Christian Giske research group
- Tissue immunology – Research group Helen Kaipe
- Health Economics and Economic Evaluation – Niklas Zethraeus' group
- Mental Health Neural Dynamics Lab – Kristoffer Månsson's research group
- Stem Cell Biology – Jonas Muhr's Group
- VIVAC research group: Vaccines and Immunotherapies against Viruses And Cancer – Matti Sällberg
- The passage of the mRNP particle through the nuclear pore – Bertil Daneholt's research
- Therapeutic Immunology and Transfusion Medicine – Michael Uhlin’s research group
- Musculoskeletal conditions and sports medicine research – Iben Axén's research group
- Research groups in cancer and oncology
- Nutritional neuroscience – Janina Seubert's Research Group
- Tobias Karlsson's group
- Research groups in pharmacology and toxicology
- Precision Psychiatry – Lu Yi's research group
- Research groups in medicinal chemistry
- Agneta Richter-Dahlfors group
- Cancer epidemiology with focus on breast cancer and applied biostatistics – Anna Johansson's research group
- Autism, ADHD and other neurodevelopmental conditions – Sven Bölte's research group
- Twinstudies on health conditions and sickness absence – Pia Svedberg's research group
- Psychiatric comorbidity and intergenerational transmission of mental health problems – Erik Pettersson's research group
- Genetic and pharmacological epidemiology – Zeberg laboratory
- REACH – digital treatment and prevention – David Moulaee Conradsson's research group
- Mucosal immunology lab – Charlotte Thålin research group
- Stroke – Annika Lundströms research group
- Loh
- Gonçalo Castelo-Branco Group
- Pediatric Neuroimmunology and Epilepsy – Ronny Wickström's research group
- Explores various cell signaling phenomena and their impact on critical biological and medical processes using advanced light microscopy – Per Uhlén Group
- Neural stem cells – Anna Falk group
- Diabetes and Associated Complications – Sergiu-Bogdan Catrina's research group
- Myelodysplastic syndromes – Eva Hellström Lindberg group
- Immune responses to human viral infections and cancer – Hans-Gustaf Ljunggren group
- Health services research in neurological conditions – Charlotte Ytterberg's research group
- Neurosurgery – Mikael A. Svensson's research group
- Westman neuroimaging group
- Stemcells and Inflammation – Lou Brundin's research group
- Pharmacological nitric oxide research – Jon Lundberg's research group
- Epidemiology of Psychiatric Conditions, Substance use and Social Environment (EPiCSS) – Emilie Agardh's & Renee Gardner's Research Group
- Autoimmune neurology – Jakob Theorell team
- Reducing antibiotic resistance – Research group Pontus Nauclér
- Orthopaedics – Karl Eriksson's research group
- Endocrine Surgery – Robert Bränström's research group
- Paediatrics – Inger Kull's research team
- Cancer proteogenomics - from methods to clinical applications – Janne Lehtiö's Group
- Primary Immunodeficiency, Innate Immunity and Antimicrobial peptides – Peter Bergman Research Group/The AMP-group
- Clinical Virology and Immunology – Annika Karlsson's research group
- Jussi Taipale Group
- Global Child Health and the Sustainable Development Goals – Tobias Alfvén's research group
- Topoisomerases, chromatin biology and cancer – Laura Baranello's group
- Stem cells and neural development – Johan Ericson's Group
- Vascular morphogenesis and function in health and disease – Lars Jakobsson group
- Epidemiology and Public Health Intervention Research (EPHIR) – Jette Möller's research group
- Epithelial stem cells in development and disease – Maria Genander's group
- Research Group Gustaf Edgren
- Drug discovery, pancreatic beta-cell regeneration – Olov Andersson's Group
- Unit of Biostatistics – Matteo Bottai
- Research groups in cardiology and cardiovascular diseases
- Sleep, cognition and health – John Axelsson's research group
- Research groups in infectious medicine
- Research groups in physiology and anatomy
- Cardiometabolic epidemiology and aging – Ida Karlsson's research group
- How the overview of research fields is made
- Precision Cancer Control Group – Martin Eklund's research group
- Liver and monocyte remodelling in non-alcoholic fatty liver disease and cardiovascular disease – Rongrong Fan's research group
- Early childhood development and neurodevelopmental conditions – Terje Falck-Ytter's research group
- Mental health and social integration (Mente) – Ellenor Mittendorfer-Rutz's research group
- Mechanisms of protein aggregation and inhibition – Axel Abelein group
- Accelerating drug discovery using molecular modeling and machine learning – Andreas Luttens' group
- Pediatric Healthcare Science – Cecilia Bartholdson research group
- Digital Strategies for AF Detection and Outcome Prevention – Emma Svennberg Team
- Speech-Language Pathology – Liza Bergström's research group
- Arrhythmia – Johan Engdahl's research group
- Environment, nutrition and health – Anna Bergström's research group
- Genetic and epigenetic factors in asthma and allergy – Cilla Söderhäll's research group
- Cardiovascular epidemiology – Team Bruna Gigante
- Neurobiology of Stress and Treatment Response – Juan Pablo Lopez group
- Vascular Surgery – Ulf Hedin's research group
- Annika Bergquist group
- Acute myeloid leukemia – Sören Lehmann group
- Human tissue-resident NK cells – Niklas Björkström group
- Neuropsychoimmunology – Sophie Erhardt's research group
- Diabetes epidemiology – Sofia Carlsson's research group
- Stroke - Acute Intervention and Secondary Prevention – Niaz Ahmed's research group
- Molecular muscle physiology and pathophysiology – Lanner Lab
- Community nutrition and physical activity (CoNPA) – Liselotte Schäfer Elinder's research group
- Inflammatory bowel disease (IBD) – Research group Eduardo Villablanca
- CRISPR-based drug target discovery in cancer and autoimmunity – Research group Fredrik Wermeling
- Emergency care – Kristian Ängeby's research group
- Clinical Cancer Epidemiology – Research group Karin Ekström Smedby
- Cancer prevention and screening – Johannes Blom's research group
- Somatosensation – Patrik Ernfors group
- DNA replication & Cancer Genetics – Lemmens Group
- Laboratory testing for alcohol and drugs of abuse – Anders Helander research
- Anaesthesia and Intensive care – Rebecka Rubenson Wahlin/Anna Schandl's research group
- Jonas Fuxe Group
- SCF ubiquitin ligases, cell cycle, transcription and cancer development – Olle Sangfelt's Group
- Toxicological Mechanisms – Emma Wincent's research group
- Stem Cells in Tissue Homeostasis and Regenerative Medicine – Jonas Frisén's Group
- Urology – Olof Akre's research group
- The New World of Work – Theo Bodin's research group
- Midbrain dopaminergic neuron development
- Research group Michael Fored
- Neurobiology of pain & Therapeutics – Saida Hadjab Group
- Risk assessment – Mattias Öberg's research group
- Research groups in cell and molecular biology
- Developmental Psychology: Digital Media and ADHD – Lisa Thorell's research group
- Research groups in education and pedagogy
- Research groups in biostatistics and probability theory
- ESSI – Emotion regulation, Self-injury, Suicide, and Intervention – Johan Bjureberg's research group
- Medical ethics – Gert Helgesson's group
- Treatment of substance use disorders – Johan Franck's Research Group
- Sex hormones and sex differences in diseases of the brain – Ivan Nalvarte's research group
- Cilia in the brain – and their connections to human brain disorders – Peter Swoboda's research group
- Evidence-based methods in autism and ADHD – Tatja Hirvikoski's research group
- Long-Term Outcomes after Perioperative and Intensive Care – Max Bell research group
- Translational and clinical research in acute myeloid leukemia – Team Martin Jädersten
- CEREBRA – Susanne Palmcrantz's research group
- Ute Römling group
- Perioperative quality – Jan Jakobssons research group
- Translational Pharmacology – Kent Jardemark's team
- Global and Sexual Health (GloSH) – Anna Mia Ekström's research group
- Medical Inflammation Research – Rikard Holmdahl group
- Neuro-infections & Neuroinflammation – Federico Iovino Group
- Clinical Chemistry and Blood Coagulation – Jovan Antovic's research group
- Translational research on human microbial infections and its consequences – Anders Sönnerborg's group
- Human tissue-resident NK cells in homeostasis and disease – Nicole Marquardt team
- Etiology and pathogenesis of type 1 diabetes – Malin Flodström-Tullberg group
- Oxygen sensing, cancer and intratumor heterogeneity – Schlisio Lab
- Genetic Epidemiology of Neuroinflammatory Disorders – Ingrid Kockum's research group
- Retina – Anders Kvanta's research group
- Glaucoma – Pete Williams' research group
- Leadership in healthcare and academia – Mia von Knorring's group
- Luminal gastroenterology – Research group Charlotte Hedin
- Identifying molecular signals in the genital mucosa that determine susceptibility to sexually transmitted infections – Research group Kristina Broliden
- Coronary heart disease – Research group Per Svensson
- Educational development – Matti Nikkola's group
- Post-transcriptional regulation in mitochondria – Joanna Rorbach group
- Cellular diversity is generated during development, neuronal identity – Johan Holmberg's Group
- Lipid handling in health and cardiometabolic disease – Research group Carolina Hagberg
- Renal glomerulus biology and diseases – Jaakko Patrakka group
- Research group Caroline Palm Apergi
- Precision pathology and tumor heterogeneity – Johan Hartman's Group
- Lifelong Learning in Health Care Contexts – Terese Stenfors' group
- Translational Arthritis Research – Team Bence Réthi
- Stem cells, Developmental Biology, Heart Disease, Heart Development, Genetics, Biotechnology – Kenneth R. Chien's group
- Urological cancer – Lars Egevad's Group
- The ubiquitin/proteasome system in neurodegenerative diseases and cancer – Nico Dantuma's Group
- Experimental alcohol- and drug dependence research – Vladana Vukojević research group
- Chronic inflammatory disease epidemiology – Research group Johan Askling
- Statistical and bioinformatics analyses of high-throughput molecular data – Yudi Pawitan's research group
- Urban environment and children´s and youth health – Olena Gruzieva's research group
- Research groups in laboratory medicine
- Research groups in medical biotechnology
- Research groups in physiotherapy
- Breast cancer epidemiology – Kamila Czene's research group
- Research groups in human computer interaction
- Research group Liv Eidsmo
- Preventive Medicine – Susanna Larsson's research group
- Transition to adulthood for individuals with neurodevelopmental conditions – Ulf Jonsson's team
- Function and Health in Respiratory and Cardiovascular Conditions – Malin Nygren-Bonnier's research group
- Laboratory of lymphocyte biology – Team Taras Kreslavskiy
- Global Reproductive Outreach & Wellness (GROW) – Michael Wells
- Mats Marshall Heyman research group
- SLE/APS/Vasculitis group – Research group Aleksandra Antovic
- Itziar Martinez Gonzalez group
- CoNFIND Cognition and Fatigue – Marika Möllers research group
- Tissue immunosurveillance by cytotoxic lymphocytes – Takuya Sekine team
- Physical activity and Sports medicine with focus on prevention – Hagströmer research group
- Receptor biology and signaling – Schulte Lab
- Neurobiology of Obesity – Alessandro Furlan group
- Upper GI Surgery – Jesper Lagergren's research group
- Sepsis and COVID-19 – Kristoffer Strålin Group
- Harnessing Neutrophils for Precision Cell Therapy against solid tumors – Roland Fiskesund Team
- Human T cell immunity to evolving viruses and cancers – Marcus Buggert group
- Neuroradiology - MRI physics – Stefan Skare's research group
- Wilhelm lab
- Inflammatory responses in cancer and autoimmunity – Mikael Karlsson's Group
- Cultural Medicine – Solvig Ekblad's unit
- Working life, ergonomics, psychosocial factors, and health – Research group Daniel Falkstedt
- Aging and health – Welmer's research group
- NK Cell Recognition – Klas Kärre Group
- Computational Breast Imaging – Fredrik Strand's Group
- Drug treatment – Erik Eliasson research group
- Neural differentiation as a strategy for neuroblastoma treatment – Marie Arsenian Henriksson Group
- Cancer immunotherapy – Mattias Carlsten group
- Cell-based immune therapy for cancer – Andreas Lundqvist's Group
- Basic molecular structure of human skin – Lars Norlén's research
- Reproductive, Perinatal and Pediatric Epidemiology – Research group Olof Stephansson
- Research group Erik Melén
- Research group Joel Nordin
- MINT – Klas Karlgren's team
- Translational breast cancer research – Theodoros Foukakis' Group
- Genes function and molecular bases of diseases – Research group Magdalena Paolino
- Homeostasis and tissue repair mechanisms in the mammalian system, pericytes, stem cells – Christian Göritz Group
- The Perinatal Epidemiology Lifecourse Lab – Team Neda Razaz
- Experimental Cancer Medicine (ECM)
- Research group Therese Djärv
- Molecular and cellular neuroendocrinology – Tibor Harkany group
- Neuropsychiatric disorders – Håkan Karlsson group
- Research groups in dermatology and venereal diseases
- Evidence-based practice: prevention, intervention and implementation – Pia Enebrink's research group
- Research groups in radiology and medical imaging
- Research groups in psychiatry
- Maria Eriksdotter research group
- Research groups in precision medicine
- Breast cancer surgery – Jana de Boniface's research group
- ICare – Ann Langius-Eklöf's research group
- Immunology and Chronic Disease – Johan Frostegård's research group
- Health in Everyday Life (HELD) – Susanne Guidetti's research group
- Center for Resuscitation Science – Jacob Hollenberg's research group
- Applied Developmental Neurobiology – Carl Sellgren research group
- Reproductive Health/Reproductive Medicine – Kristina Gemzell Danielsson's research group
- Malin Holzmann's research group
- Radiation therapy/ Radiation Oncology – Pehr Lind Group
- Fossati
- APP processing and Abeta localization at super-resolution and glycan biomarkers for Alzheimer disease – Sophia Schedin Weiss's research group
- ALGOSH: Algorithmic Management at Work
- Development of autonomic control – Herlenius Research
- Maintenance and expression of mtDNA in disease and ageing – Nils-Göran Larsson Group
- Neuropharmacology - Movement Disorders – Per Svenningsson's research group
- Somatosensation & Gargalesis – Konstantina Kilteni group
- Signal Transduction – Ismael Valladolid Acebes' research group
- Microbiome and Infections – Piotr Nowak Research Group
- Immunological tolerance and transfusion immunology – Petter Höglund group
- Immunopathogenesis and new therapeutic strategies in tuberculosis – Susanna Brighenti group
- Pain and Brain Imaging – Karin Jensen's research group
- Susanne Nylén group
- Hand surgery – Maria Wilckes Research group
- Environmental impact on pulmonary host defence and chronic airflow obstruction – Anders Lindén's research group
- Rehabilitation, collaboration and aging – Elisabeth Rydwik's research group
- Tuberculosis Aerobiology – Antonio Rothfuchs group
- Oscar Fernandez-Capetillo group
- Diversity within the T cell response, T cell memory – Research group Carmen Gerlach
- Clinical and translational melanoma research – Hanna Eriksson's Group
- Meiotic chromosome segregation – Christer Höög's research group
- B cells and autoantibodies in rheumatic disease – Team Caroline Grönwall
- Precision medicine in lymphoid malignancies including CLL; from new targets to real-world assessments – Anders Österborg's Group
- Research group for Cancer evolution
- Jiri Bartek group
- Translational Cardiology (@TransCardio) – Research group Magnus Bäck
- Gene regulation, genome-wide experimental and computational techniques – Rickard Sandberg's Group
- The Helleday Laboratory focuses on harnessing defects in the DNA damage response and metabolism to develop novel therapies
- Small RNAs in cancer development – Weng-Onn Lui's Team
- Geriatric pharmacoepidemiology – Kristina Johnell's research group
- Molecular neurodevelopment and neuro-oncology – Ola Hermanson group
- Research groups in anesthesiology and intensive care
- Gastrointestinal epidemiology – Jonas Ludvigsson's research group
- Health inequalities and minority stress – Richard Bränström's research group
- Research groups in microbiology
- Research groups in orthopaedics
- Hereditary hematological malignancies – Bianca Tesi team
- Lars Holmgren's Group
- Substance use prevention – Johanna Gripenberg's research group
- Perioperative care – Ulrica Nilsson's research group
- Mechanisms behind HIV-1 latency and rebound – Peter Svensson's group
- ENGAGE- Enacting health and change through everyday activity – Patomella group
- Diagnosis and treatment of children with asthma and allergy: from molecular diagnosis to intervention – Jon Konradsen's research group
- Translational research in childhood cancer and histiocytic diseases – Nikolas Herold Research group
- Coronary artery disease – research at Danderyd hospital
- Mechanisms of malignant hematopoiesis – Pedro Moura team
- Regulation of B cell activation – Pia Dosenovic Group
- Complex interventions – Team Carina King
- Reproductive endocrinology and metabolism – Elisabet Stener-Victorin's research group
- Tissue engineering – Magdalena Fossum team
- Inflammation and metabolism – Nicolas Pillon's team
- Neurobiology of Sensory Systems – François Lallemend group
- Experimental Traumatology Research Unit
- Genetic Modification of NK cells for Optimized Functions against Cancer – Arnika Wagner team
- NK cells in the development of adaptive immune responses – Benedict Chambers team
- Molecular pain research – Camilla Svensson's research group
- Brain Connectomics – Joana Pereira's research group
- Genomic Science and RNA Biology – Vicent Pelechano group
- Immunotherapy – Robert Harris' research group
- Infectious diseases "Venhälsan" – Jaran Eriksen's research group
- Musculoskeletal disorders from a biopsychosocial perspective – Wim Grooten's research group
- Hjerling-Leffler group
- Respiratory and invasive infections and microbial pathogenesis – Birgitta Henriques-Normark Laboratory
- Multimodal Brain Imaging – Daniel Lundqvist's research group
- Obsessive - Compulsive and Related Disorders Across the Lifespan – David Mataix-Cols research group
- With a focus on proteins for better cancer treatments – Pär Nordlund's Group
- Precision cancer medicine and precision radiotherapy in lung cancer - from molecular mechanisms/biomarkers to clinical trials – Kristina Viktorsson's team
- Cancer Bioinformatics – Nick Tobin's Team
- Lipoproteins and the Immune System – Research group Stephen Malin
- Equity and Health Policy (EHP) – Ann Liljas' research group
- Effects of hypoxia in physiological and pathological contexts, Hypoxia Inducible Factors (HIF) – Randall Johnson's Group
- Chemistry II – Jesper Z. Haeggström group
- Genomic analysis of parasites and viruses, metagenomic sequencing – Björn Andersson's Group
- Translational control of cancer – Ola Larsson's Group
- Gastrointestinal Surgery KI SÖS – Gabriel Sandblom's research group
- Understanding the mechanisms behind hantavirus-mediated pathogenesis – Jonas Klingström group
- The Gynaecological research team – Elisabeth Epstein
- Understanding how life develops – Emma Andersson's Group
- Statistical methods in epidemiology – Paul Dickman's research group
- Neurons and Neural Networks – Ole Kiehn group
- Research groups in developmental biology
- Behavioral, Resilience and Digital Health in Pain – Rikard Wicksell’s research group
- Research groups in neurology
- Research groups in epidemiology, global public health and social medicine
- Nordic Brain Network – Miia Kivipelto's research group
- Resilience and mental health – Serhiy Dekhtyar group
- Forensic behavioral and neuroscience – Katarina Howner's Research Group
- Immune Engineering – Team Leo Hanke
- Chemical and physical exposure in the work environment and health – Jenny Selander's research group
- Mechanobiology of cardiac regeneration – Elif Eroglu's Group
- Tumors of the female genital organs – Miriam Mints' research group
- Psychoneuroimmunology – Mats Lekander's research group
- Research groups in odontology
- Neuroimmunovascular biology – Harald Lund's team
- Improved synthetic DNA structures by evolutionary selection – Erik Benson Group
- Lifestyle and Kidney Cancer – Stephanie Bonn Team
- Neuroplasticity and Regeneration – Konstantinos Ampatzis group
- Paediatric cell and molecular biology – Hjalmar Brismar's research group
- Quantitative Biology of the Nucleus – Magda Bienko Group
- Cardiometabolic factors, brain aging, and dementia care – Weili Xu group
- Cognitive Neuroscience of Body and Self – Henrik Ehrsson Group
- Surgical Care Science – Pernilla Lagergren's research group
- Clinical and translational studies on viral hepatitis – Soo Aleman group
- Cell and Gene Therapy – Evren Alici group
- Personalized Medicine and Drug Development – Lauschke-lab
- Electrophysiological neuropharmacology – Göran Engberg's research group
- Mechanisms of Pain and Treatment – Eva Kosek's research group
- Functional (Epi)genomics of Neuroinflammatory Diseases – Maja Jagodic's research group
- Molecular basis of gene regulation of diseases – Carsten Daub's research group
- Health risk assessment methodology – Anna Beronius' group
- Medical genetics – Catharina Larsson's Group
- Translational Genetics of Neurodegenerative disease – Caroline Graff's research group
- Development, transcription factors, neurons, dopamine – Thomas Perlmann's Group
- Clinical and translational research within lupus and autoimmunity – Team Ioannis Parodis
- Cell cycle, mitotic entry, and DNA-damage checkpoints – Arne Lindqvist's Group
- Breast Surgery – Irma Fredriksson's research group
- Laura Orellana's Group
- Research group Ali Mirazimi
- Early drug discovery research in chronic inflammatory diseases – Research group Michael Sundström
- Integrative Physiology – Juleen Zierath's research group
- Simon Elsässer group
- Clinical Neuroimmunology and Immunomodulation – Fredrik Piehl's research group
- Understanding mechanisms of vaccination – Research group Karin Loré
- Systems regenerative neurobiology – Enric Llorens Group
- Clinicial Cancer Genomics – Johan Lindberg's research group
- Jeanette Hellgren Kotaleski group
- Brain control of food intake and body weight – Björn Meister group
- Research groups in endocrinology and diabetes
- Research groups in gastroenterology and hepatology
- Research groups in neurosciences
- Research groups in respiratory medicine and allergy
- Caring in Community Care – Zarina Nahar Kabir's research group
- Molecular mapping of the nervous system in health and disease – Jan Mulder group
- Child and adolescent psychiatry – Jens Högström's research group
- Genetic mechanisms of ageing – Maria Eriksson's research group
- Digital Psychiatry – Philip Lindner's research group
- Synaesthesia, autism and perception – Janina Neufeld's team
- Vascular complications in cardiometabolic disease – Team Zhichao Zhou
- Lars Karlsson team
- Sepsis and microcirculation – Sara Tehrani's research group
- Translational Clinical Orthopaedics – Michael Axenhus Research Group
- Antigen-specific T cell repertoires – Cassotta lab
- Translational Molecular Imaging – Nordberg Lab
- Neuroplasticity – Dagliyan group
- Developmental and Translational Neurobiology – Cristiana Cruceanu's research group
- Microbiota–gut–brain axis and neurodevelopment – Rochellys Diaz Heijtz group
- Thoracic Surgery – Anders Franco-Cereceda's research group
- Electrophysiology – Mats Jensen-Urstad group
- Role of T cells in human host defense – Johan Sandberg group
- Germ cell biology and developmental programming in epigenetic inheritance of diseases – Qiaolin Deng's research group
- Unit for Bioentrepreneurship – Hanna Jansson's group
- Health informatics – Sabine Koch's group
- Cancer, rapid ageing and nutrition – Martin Bergö's research group
- Clinical radiation therapy – Åsa Carlsson Tedgren's Group
- TRANslational Theranostics Group – Thuy Tran
- Pathogenic pathways in Alzheimer Disease – Lars Tjernberg's research group
- Hematological and solid tumors – Research group Xu Dawei
- Developmental Neurogenomics – Michael Ratz's Group
- Notch signaling, ES cells, myogenic and vascular progenit breast cancer – Urban Lendahl's Group
- Particle toxicology – Hanna Karlsson's group
- Mitochondrial dysfunction in Alzheimer Disease – Maria Ankarcrona's research group
- Molecular pathology of the lung and pleura – Katalin Dobra's Group
- Late effects and cancer survivorship after diagnosis and treatment of aggressive lymphoma – Team Sandra Eloranta
- Research Group Anna Nopp
- Coding and non-coding RNAs in cancer – Per Hydbring's Team
- Research group Mikael Björnstedt
- Sister chromatid cohesion in DNA damage responses, DSB repair, Genome Integrity, Gene regulation – Lena Ström's Group
- Inborn Errors of Endocrinology and Metabolism – Anna Wedell's research group
- Functional Precision Medicine – Brinton Seashore-Ludlow's Team
- Understand skin using modern biology – Maria Kasper's Group
- Integrative Epidemiology – Fang Fang's research group
- Molecular Biometry – Roman Zubarev group
- Idiopathic pulmonary fibrosis (IPF) – Research group Magnus Sköld
- Molecular Neuroscience – Carlos Ibáñez group
- Research groups in psychology
- Research groups in forensic science
- Research groups in medical genetics and genomics
- Research groups in nursing
- Research groups in rheumatology and autoimmunity
- Genetic epidemiology of prostate and testicular cancer – Fredrik Wiklund's research group
- Systems biology of aging – Juulia Jylhävä's research group
- Aging – basic mechanisms and interventions – Christian Riedel's research group
- Embryonal, foetal and brain development – Juha Kere's research group
- Research groups in occupational therapy
- Growth and Cartilage Biology – Ola Nilsson's research group
- Infections and immunity in children with cancer – Anna Nilsson's research group
- Oxygenation, early resuscitation in trauma and drug treatment in cardiac arrest – Team Malin Jonsson Fagerlund
- Prehospital Emergency Care – Veronica Vicente's Research Group
- The brain after surgery and trauma - neuroinflammation, cognitive impairment and dementia – Lars I Eriksson research group
- Personalized diet and medications for cognition – Garcia-Ptacek group
- Arthroplasty – Olof Sköldenberg’s research group
- Anna Undeman Asarnoj team
- Visual Neuroscience – Rune Brautaset's research group
- Chemicals and female fertility – Pauliina Damdimopoulou Research Group
- Molecular and cellular exercise physiology – The Ruas Lab
- Neural circuits of cognition – Marie Carlén group
- Colorectal Surgery – Anna Martling and Caroline Nordenvall's research group
- Myeloma group – Evren Alici's research group
- Medicine and history of science – Eva Åhrén's group
- Cardiometabolic diseases – Ping Chen research group
- Anders Mutvei research group
- Ocular Oncology and Pathology – Gustav Stålhammar's research group
- Clinical Neurophysiology – Charith Cooray's research group
- Regulation of Gene Expression during Viral Infection – Gerald McInerney Group
- Social and affective learning and decision-making – Andreas Olsson's research group
- Translational Breast Cancer Research: Long-Term Risk and Endocrine Treatment Benefit – Linda Lindström's Group
- Notch3 and the cerebral small vessel disease CADASIL – from molecular mechanisms to treatment strategies – Helena Karlström's research group
- Dementia and Multimorbidity – Dorota Religa's research group
- Diagnostic Radiology – Lennart Nedar's research group
- Cognitive Neuropsychiatry – Predrag Petrovic's research group
- Chemical carcinogenesis – Ulla Stenius' group
- Metal toxicology – Maria Kippler's research group
- Cellular Stress in Health and Disease – Federico Pietrocola's group
- Autophagy and cardiovascular disease – Team Ewa Ehrenborg
- Research group Birgitta Sander & Birger Christensson
- Innate Immune Regulation – Quirin Hammer team
- Genetics of tumor predisposition and progression in breast cancer – Svetlana Bajalica Lagercrantz's Team
- Implementation and Quality (IMPAQT) – Claudia Hanson's research group
- Structural and Biophysical Immunology – Research group Adnane Achour
- Ulrika Marklund group
- Sustainable work & occupational safety and health – Emma Brulin's research group
- Health Systems Leadership, Management, and Safety – Carl Savage's group
- Cell and Molecular Immunology - Research group Cecilia Söderberg Naucler
- Brain Circuits – Konstantinos Meletis group
- Petter Ljungman's research group- Health effects of the Ambient Environment
- Neuropsychology of music – Fredrik Ullén group
- Research groups in geriatrics and gerontology
- Research groups in nutrition and dietetics
- Research groups in sports and fitness sciences
- Prostate Cancer – Henrik Grönberg's research group
- Methods of health promotion in the workplace – Lydia Kwak's research group
- Autism and perinatal epidemiology – Sven Sandin research group
- Wengström's research group
- Modelling and simulation with applications to cancer epidemiology and health economics – Mark Clements' research group
- Health Technology Assessment – Monica Hultcrantz’s group
- Pediatric anesthesiology Stockholm – Per-Arne Lönnqvist's research group
- Biophysics – Erdinc Sezgin's Lab
- War, crisis, and security studies
- Systems Medicine – Johan Björkegren research group
- Trauma – Max Gordon's research group
- Perpetration Prevention – Rahm and Joleby's research group
- SOLIID - Sustainable Organizational Learning, Innovation, Improvement and Development in Health and Social services – Monica Nyström's group
- Social Gerontology – Carin Lennartsson group
- Genetic and molecular basis of nervous system disorders – Andrea Carmine Belin group
- Chronobiology – Petrus lab
- Leukemic stem cells – Petter Woll group
- Nanomedicine and Spatial Biology – Teixeira Lab
- Global Disaster Medicine - Health Needs and Response – Johan von Schreeb's research group
- Translational cardiac and skeletal muscle physiology – Daniel Andersson's research group
- Neuroradiology – Staffan Holmin's research group
- Tumour genomics – Lauri Aaltonen's research group
- Neurobiology of Motor Actions – Abdel El Manira group
- Neuroinflammation in Alzheimer disease – Marianne Schultzberg research group
- Palliative Medicin – Linda Björkhem-Bergman's research group
- Nucleotide metabolism and molecular pharmacology – Sean Rudd's Group
- Cardiovascular Inflammation – Research group Peder Olofsson
- Mesenchymal stem cells – Katarina Le Blanc group
- Luigi De Petris' Team
- Autoimmunity and cancer – Research group Marie Wahren-Herlenius
- Atherothrombosis research – Team Nailin Li
- Precision cancer medicine – Olli Kallioniemi's group
- Research group Stephan Mielke
- Genetics of autoimmune diseases – Team Leonid Padyukov
- Musculoskeletal disorders; planetary health & biopsychosocial approaches – Nina Brodin's research group
- Vascular Functions in Metabolic and CNS Diseases – Ulf Eriksson group
- Sarcoma Genomics – Felix Haglund de Flon's Group
- Team Helene Alexanderson
- Injury and Repair in the Immature Brain – Blomgren group
- Statistical methods for cancer patient survival – Therese Andersson's research group
- Individual differences in cognition – Agneta Herlitz’ research group
- Research groups in health economy, healthcare management and policy
- Research groups in reproductive medicine and gynaecology
- Research groups in structural biology
- Research groups in artificial intelligence
- Work environment, health and productivity – Christina Björklund's research group
- Predictive medicine – Mattias Rantalainen's research group
- Epigenetic regulation of leukemia and normal blood development – Andreas Lennartsson's research group
- Epigenetic mechanisms underlying metabolic-inflammatory diseases – Eckardt Treuter's research group
- The IMPACT research group – Marie Löf
- Stockholm Center for Health Economics (StoCHE) – Emelie Heintz's team
- Towards personalized physiology in Intensive Care – Team Johan Mårtensson
- Ninib Baryawno research group
- Neurohormonal Basis of Physiological and Maladaptive Expressions of Survival Behaviors – Stefanos Stagkourakis group
- Breast cancer precision medicine – Marike Gabrielson's research group
- D-WHIN: Danderyd Wrist and Hand INitiative – Research group
- Research group Kilian Eyerich
- Global infections – Research group Anna Färnert
- Translational Microbiome Research and Pandemic Preparedness – Lars Engstrand group
- EcoMind and biological pathways in cognitive aging – Debora Rizzuto group
- Circuits controlling Action and their Evolution – Sten Grillner group
- Mikael Rydén & Niklas Mejhert lab
- Understand Fundamental Immune Mechanisms to Develop New Treatments for Lung Diseases – Tim Willinger group
- Clinical Brain Imaging Methods – Anna Falk Delgado's research group
- Hyperbaric medicine – Peter Lindholm's research group
- Eye movements and vision – Tony Pansell's research group
- Lung toxicology – Lena Palmberg's research group
- The NeuroCardioMetabol Group – Thomas Nyström & Cesare Patrone
- Environmental and genetic factors influence on neurodevelopment – Sandra Ceccatelli group
- Skin wound healing – Research group Ning Xu Landén
- From development to disease: decoding stem cell programs for CNS repair – Erik Sundström's and Xiaofei Li's research group – Erik Sundström-Xiaofei Li group
- Neurodegenerative diseases and Aging – Daniel Ferreira Research Group
- Deep learning models of cancer mechanisms – Avlant Nilsson's Group
- Clinical Physiology – Marcus Carlsson's research group
- Molly Stevens group
- Molecular Mechanisms of Viral Oncogenesis – Maria G. Masucci's Group
- Björn Reinius group
- Quantitative principles in tumour biology – Jean Hausser's Group
- Bone biology – Sara Windahl Research group
- Research group Volkan Özenci
- Genetics and mechanisms of metals – Karin Broberg's research group
- Immunodeficiency Diseases – Lisa Westerberg Group
- Structural studies of fertilisation and zona pellucida module proteins – Luca Jovine's research group
- Tissue regeneration, focusing on fat cells (adipocytes) and obesity – Kirsty Spalding's Group
- Robert Månsson group
- Developmental and Regenerative Stem Cell Medicine – Lanner Lab
- Suicide and mental health lab – Vladimir Carli and Gergö Hadlaczky's group
- Medical, genetic and molecular decoding of neurodevelopmental disorders – Kristiina Tammimies' research group
- Working hours, Recovery and Safety in working life. – Anna Dahlgren's research group
- Research groups in hematology
- SOMIND: Somatic, Mental, and Neurodevelopmental Conditions – Agnieszka Butwicka's research group
- Research groups in drug abuse and addiction
- Research groups in biomaterials science
- Mikael Adner's research group
- Chemical and functional neuroanatomy of pain and stress systems – Tomas Hökfelt group
- Spider silk biology for biomedical applications – Anna Rising's research group
- Cancer Epidemiology and Causal Inference – Research group Maria Feychting
- Protein biochemistry for development of novel medical treatments – Janne Johansson's research group
- Healthcare financing and organization – Clas Rehnberg's group
- Space and environmental physiology – Rodrigo Fernandez-Gonzalo group
- Novel treatment approaches for neuroblastoma – Malin Wickström Näsman research group
- Cancer immune and gene therapy – Rolf Kiessling's Team
- Pancreas biology – Amanda Andersson Rolf's Group
- CAIR-LAB: Clinical AI Research Laboratory – group
- Atopic dermatitis – Research group Emma Johansson
- Reproductive endocrinology and metabolism – Angelica Lindén Hirschberg's research group
- Genetic basis for B and T cell recognition and function – Gunilla Karlsson Hedestam Group
- Cognitive Aging and Mental Health – Erika Jonsson Laukka Group
- Molecular and circuit neuropharmacology – Gilberto Fisone group
- The role of immunity in shaping adipose tissue development and metabolism – Jurga Laurencikiene group
- Chronic myeloid malignancies – Johanna Ungerstedt group
- Studies of tissue microbiology and immunology – Mattias Svensson group
- Sten Linnarsson group
- Balance, gait, exercise and physical activity in neurological diseases – Franzén Group
- Global Health Pharmacology and Therapeutics (GH-Pharma) – Eleni Aklillu's research group
- Real-world-data in Multiple Sclerosis – Anna Glaser's research group
- Suicidology and transcultural psychiatry – Marie Dahlin's research group
- Experimental and Clinical Neuroendocrinology – Maria Petersson's research group
- Cutting edge translational research of common and rare skin disease – Research group Jakob Wikström
- Alzheimer’s Disease Risk Factors & Sex-Specific Mechanisms – Silvia Maioli's research group
- Frontal lobes and cognitive failure – Wahlund's research group
- Clinical Pharmacology – Research group Rickard Malmström
- Molecular Vascular Medicine – Team Lars Maegdefessel
- Molecular endocrine pathology – Christofer Juhlin's Group
- Presynaptic mechanisms – Lennart Brodin group
- Malignant melanoma – Hildur Helgadottir's Group
- Precision strategies to eradicate heterogeneous tumor cells in pediatric cancer – Kasper Karlsson's Group
- Respiratory and systemic immune responses in human pulmonary viral infection and inflammation – Research group Anna Smed Sörensen
- Genetic toxicology – Kristian Dreij's group
- Identifying new molecular targets for the treatment of motor neuron diseases – Eva Hedlund's research
- Epigenetics - basic mechanisms and disease – Karl Ekwall's research group
- Forensic medicine – Henrik Druid's Group
- Ageing and Health – Karin Modig's research group
- Perception Neuroscience – Johan Lundström's research group
- Computational medicine and bioinformatics – Trung Nghia Vu's research group
- Toxicology – Bertrand Joseph´s Research group
- Etiology, prevention and treatment of eating disorders – Ata Ghaderi's research group
- Research groups in medical ethics and history of science
- Research groups in occupational and environmental health
- Research groups in surgery
- Research groups in biophysics
- Research Group Johan Ärnlöv
- Narrative in health and social care – Research group
- Cellular and molecular mechanisms underlying intervertebral disk formation and Diversity and plasticity of adipose tissue cell niche – Meng Xie's research team
- Neuroimmunology of cancer – Sébastien Talbot
- Pediatric Systems Immunology – Petter Brodin's research group
- Microvascular oxidative stress in the triad of cardiovascular, metabolic and renal disease – Mattias Carlström group
- Cardio-renal epidemiology – Juan-Jesus Carrero's research group
- Human Translational Genetics – Stefano Romeo Group
- Clinical Pancreatology – Miroslav Vujasinovic research group
- Hemostasis and Thrombosis – Mika Skeppholms research group
- Movement, Activity and Health – Eva Broström's research group
- Multidimensional health in old age – Amaia Calderón-Larrañaga group
- Molecular epidemiology of aging – Sara Hägg's research group
- Long noncoding RNA in the adipocyte – Alastair Kerr Research Group
- Leukemia niche – Hong Qian group
- Disease mechanisms in severe acute bacterial infections – Anna Norrby-Teglund group
- Gastrointestinal pediatric surgery – Tomas Wester's research group
- Developmental biology and regenerative medicine – Igor Adameyko's research group
- Experimental audiology – Barbara Canlon's research group
- Translational Immunology in Neuroinflammation – Olivia Thomas' research group
- Sickness absence, health and living conditions – Emilie Friberg's research group
- Health Systems and Policy – Cecilia Stålsby Lundborg's research group
- Endocrinology and autoimmune diseases – Research group Sophie Bensing
- Health economics of Alzheimer disease and other dementias – Linus Jönsson Group
- Epidemiology: Diet, contaminants and health – Agneta Åkesson's research group
- Neuronal circuits of anxiety – Janos Fuzik group
- Paediatric rheumatology – Research group Helena Erlandsson Harris
- Cardiovascular Epidemiology – Karin Leander research group
- Molecular cardiology – Research group Francesco Cosentino
- Björn Högberg group
- Regeneration mechanisms – Andras Simon's Group
- Assessing and managing chemical risks – Linda Schenk's research group
- Autoimmunity and adaptive immunity in rheumatic disease – Research group Vivianne Malmström
- Rheumatic systemic diseases – Team Marie Holmqvist
- Heart failure with reduced and preserved ejection fraction. Clinical and translational aspects. – Research group Lars Lund
- Christoph Ziegenhain Group
- Psychiatric epidemiology – Paul Lichtenstein's research group
- Structural basis of RNA biogenesis – Martin Hällberg's research group
- Hematopoietic Stem Cell Biology Group – Sten Eirik W. Jacobsen Lab
- Psychiatric Genetic Epidemiology – Sarah Bergen's research group
- Renal Medicine at Novum (Baxter Novum) – Bengt Lindholm Group
- Research groups in biochemistry
- Psychological treatment: internet and exposure – Brjánn Ljótsson's research group
- Research groups in immunology
- Research groups in ophthalmology
- Research groups in urology and nephrology
- Research groups in sociology and migration
- Integrative physiology – Juleen Zierath and Anna Krook
- Hormonal signalling and cancer – Cecilia Williams research group
- Cell Biology of Cancer – Staffan Strömblad's research group
- Chronic inflammation with focus on rheumatology and cancer – Research group Per-Johan Jakobsson
- Health services and social work research – Lena Von Koch's research group
- Nitrate-nitrite-NO pathway in health and disease – Eddie Weitzberg's research group
- Medical sensors and diagnostics – Team Onur Parlak
- Uncovering the molecular and physical principles governing early embryonic division and nuclear organization – Jan Ellenberg Group
- Nanoscale lipid flux architecture – Veijo Salo’s Research Group
- Residual conditions after polio and neuromuscular diseases – Eva Melin's research group
Personalized Medicine and Drug Development – Lauschke-lab
We integrate 3D cell culture systems of primary human cells, microfluidics and comprehensive molecular profiling technologies to discover novel therapeutic strategies for inflammatory conditions (NASH), infectious diseases (COVID-19 and hemorrhagic fevers) and complex metabolic diseases (type 2 diabetes).
Our Research
We integrate 3D cell culture systems of primary human cells, microfluidics and comprehensive molecular profiling technologies to discover novel therapeutic strategies for inflammatory conditions (NASH), infectious diseases (COVID-19 and hemorrhagic fevers) and complex metabolic diseases (type 2 diabetes).
In addition, we use population-scale genetics and machine learning tools to map the ethnogeographic variability in genes involved in drug pharmacokinetics and pharmacodynamics with the goal to improve personalized medicine and precision public health.
Background
The number of successful drug development projects has stagnated for decades, despite major breakthroughs in chemistry, molecular biology and genetics. Unreliable target identification and poor translatability of preclinical results have been identified as major causes of failure. To improve predictions of clinical efficacy and safety, interest has shifted from conventional 2D cultures to organotypic and microphysiological culture methods in which human cells can retain physiologically and functionally relevant phenotypes for extended periods of time. The Lauschke lab develops 3D cell culture models of primary human tissues and microfluidic devices for the discovery and development. By interfacing these systems with high-throughput screening, we aim to develop novel drug candidates for a variety of indications, including NASH, type 2 diabetes, COVID-19 and hemorrhagic fevers.
In addition, we integrate population-scale genomics with machine learning to optimize pharmacogenomic drug response predictions and facilitate the implementation of NGS in personalized medicine and precision public health.
Projects
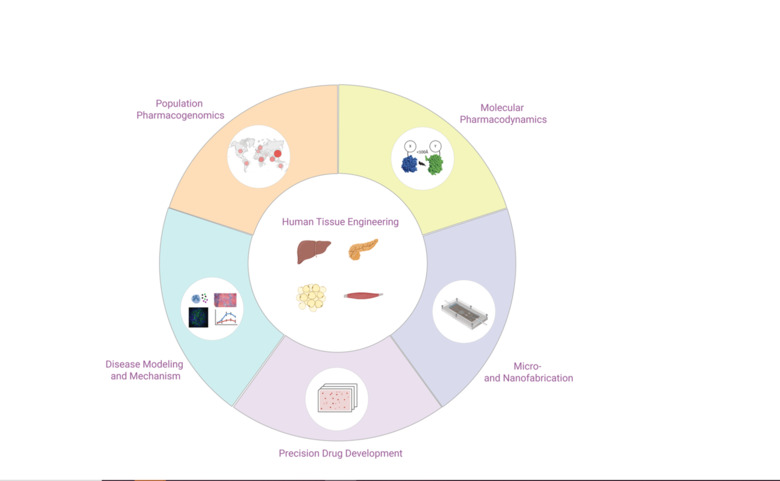
We develop and characterize long-term stable 3D human tissue models of liver, pancreas, adipose tissue and skeletal muscle. Importantly, we exclusively use patient-derived cells, i.e. no cell lines or stem cells, and integration of histological, transcriptomic, proteomic and metabolomic signatures demonstrates that the tissue models feature mature phenotypes that facilitate improved result translation. A main area of our interests is the integration of different cell types (endothelial cells, immune cells, etc.) and the functional characterization of cell-cell interactions in both health and disease.
Using state-of-the-art cleanroom tools, we leverage nanofabrication technologies for their implementation in biomedicine. Uniquely, we have established a novel nanofabrication platform, termed Nano Reaction Injection Molding (NanoRIM) for the rapid, versatile and cost-effective fabrication of polymer micro- and nanodevices. We further utilize micro- and nanopatterned surfaces to study the interaction of human cells with surface topographies.
We employ diverse materials, including polymers and inorganics, such as silicon and glass, with appropriate surface treatment for different biomedical applications. Specifically, we develop platforms for pharmacological and toxicological applications using materials with minimal drug absorption.
In the field of drug development and discovery, the poor predictive accuracy of preclinical models due to pronounced species differences has led to a high rate of project attrition. The co-culturing of human cells from different tissues and the subsequent tissue-tissue interaction in perfused microfluidic devices have shown great potential to tackle this problem in ex vivo.
By integrating our custom-designed microfluidic systems with 3D human tissue models, we have developed microphysiological organ-on-a-chip platforms with superior function that allow to mimic tissue-tissue interactions. Specifically, we integrate human 3D tissue models of liver, pancreas, adipose tissue and skeletal muscle, and utilize this platform as a model of integrative glycemic control.
We monitor signaling of G protein-coupled receptors (GPCRs) and other signal transduction events in primary human tissue models using highly sensitive and selective bioluminescence resonance energy transfer (BRET)-based biosensors. The obtained information sheds new light on the functional selectivity and subcellular signaling of important signaling cascades without the need to overexpress the receptors. Employing such biosensor-based approaches to map signaling events in fine detail holds great promise for our understanding of drug efficacy and side effects. Combining these tools with patient-derived material will provide much needed insight into the intra- and inter-patient variability of drug action and offers the potential to develop better and more targeted therapies.
Strikingly, during spheroid aggregation stages, hepatocytes first dedifferentiate, followed by rapid redifferentiation, providing an ideal ex vivo experimental paradigm to study the full spectrum of differentiation state changes that occur in vivo during liver regeneration. Besides extending our mechanistic understanding, this finding opened possibilities for the development of therapeutic approaches as a substitute for orthotopic liver transplantations. We work on the establishment of protocols in which hepatocytes isolated from patients proliferate and, after cells sufficiently multiplied, are induced to redifferentiate into functional hepatocytes using our 3D spheroid culture system. Furthermore, we investigate the molecular mechanisms underlying liver hypertrophy and develop drug candidates that improve hepatocyte proliferation.
Genetic variants primarily in drug and metabolite transporters, phase I and phase II drug metabolizing enzymes and nuclear receptors can influence drug response by modulating drug absorption, distribution, metabolism and excretion (ADME). Importantly, while in the past decades an ever-growing arsenal of genetic variants with demonstrated impacts on human drug response has been identified in these pharmacogenes, a substantial fraction of the heritable variability in drug response remains unexplained. Rare genetic variants that only occur in very few individuals and are hence missed in genome-wide association studies have been proposed to contribute to this missing heritability.
We integrate data from recent population-wide Next-Generation Sequencing (NGS) projects to quantify the extent of genetic variability in pharmacogenes on a population level and, using an arsenal of in silico techniques, quantify the impact on hepatic metabolism and pharmacokinetics and -dynamics.
Publications
Selected publications
- Article: NUCLEIC ACIDS RESEARCH. 2025;53(D1):D1498-D1509Tremmel R; Zhou Y; Camara MD; Laarif S; Eliasson E; Lauschke VM
- Article: ADVANCED SCIENCE. 2025;12(3):e2407572Youhanna S; Kemas AM; Wright SC; Zhong Y; Klumpp B; Klein K; Motso A; Michel M; Ziegler N; Shang M; Sabatier P; Kannt A; Sheng H; Oliva-Vilarnau N; Buettner FA; Seashore-Ludlow B; Schreiner J; Windbergs M; Cornillet M; Bjorkstrom NK; Hulsmeier AJ; Hornemann T; Olsen JV; Wang Y; Gramignoli R; Sundstrom M; Lauschke VM
- Article: NATURE METABOLISM. 2024;6(7):1268-1281Kizilkaya HS; Sorensen KV; Madsen JS; Lindquist P; Douros JD; Bork-Jensen J; Berghella A; Gerlach PA; Gasbjerg LS; Mokrosinski J; Mowery SA; Knerr PJ; Finan B; Campbell JE; D'Alessio DA; Perez-Tilve D; Faas F; Mathiasen S; Rungby J; Sorensen HT; Vaag A; Nielsen JS; Holm J-C; Lauenborg J; Damm P; Pedersen O; Linneberg A; Hartmann B; Holst JJ; Hansen T; Wright SC; Lauschke VM; Grarup N; Hauser AS; Rosenkilde MM
- Article: HEPATOLOGY. 2024;79(6):1337-1351Oliva-Vilarnau N; Beusch CM; Sabatier P; Sakaraki E; Tjaden A; Graetz L; Buettner FA; Dorotea D; Nguyen M; Bergqvist F; Sundstrom Y; Mueller S; Zubarev RA; Schulte G; Tredup C; Gramignoli R; Tietge UJF; Lauschke VM
- Article: NATURE MICROBIOLOGY. 2024;9(5):1499-1512Monteil VM; Wright SC; Dyczynski M; Kellner MJ; Appelberg S; Platzer SW; Ibrahim A; Kwon H; Pittarokoilis I; Mirandola M; Michlits G; Devignot S; Elder E; Abdurahman S; Bereczky S; Bagci B; Youhanna S; Aastrup T; Lauschke VM; Salata C; Elaldi N; Weber F; Monserrat N; Hawman DW; Feldmann H; Horn M; Penninger JM; Mirazimi A
- Article: NATURE METABOLISM. 2023;5(7):1188-1203Barreby E; Strunz B; Nock S; Naudet L; Shen JX; Johansson H; Soennerborg I; Ma J; Urgard E; Pallett LJ; Hu Y; Fardellas A; Azzimato V; Vankova A; Levi L; Morgantini C; Maini MK; Stal P; Rosshart SP; Coquet JM; Nowak G; Naeslund E; Lauschke VM; Ellis E; Bjoerkstroem NK; Chen P; Aouadi M
- Article: LANCET. 2023;401(10374):347-356Swen JJ; van der Wouden CH; Manson LEN; Abdullah-Koolmees H; Blagec K; Blagus T; Boehringer S; Cambon-Thomsen A; Cecchin E; Cheung K-C; Deneer VHM; Dupui M; Ingelman-Sundberg M; Jonsson S; Joefield-Roka C; Just KS; Karlsson MO; Konta L; Koopmann R; Kriek M; Lehr T; Mitropoulou C; Rial-Sebbag E; Rollinson V; Roncato R; Samwald M; Schaeffeler E; Skokou M; Schwab M; Steinberger D; Stingl JC; Tremmel R; Turner RM; van Rhenen MH; Fajardo CLD; Dolzan V; Patrinos GP; Pirmohamed M; Sunder-Plassmann G; Toffoli G; Guchelaar H-J
- Article: ADVANCED SCIENCE. 2022;9(34):e2203368Shafagh RZ; Youhanna S; Keulen J; Shen JX; Taebnia N; Preiss LC; Klein K; Buettner FA; Bergqvist M; van der Wijngaart W; Lauschke VM
- Article: GASTROENTEROLOGY. 2021;161(6):1982-1997.e11Azzimato V; Chen P; Barreby E; Morgantini C; Levi L; Vankova A; Jager J; Sulen A; Diotallevi M; Shen JX; Miller A; Ellis E; Ryden M; Naslund E; Thorell A; Lauschke VM; Channon KM; Crabtree MJ; Haschemi A; Craige SM; Mori M; Spallotta F; Aouadi M
- Article: SCIENCE ADVANCES. 2021;7(36):eabi6856Zhou Y; Arribas GH; Turku A; Juergenson T; Mkrtchian S; Krebs K; Wang Y; Svobodova B; Milani L; Schulte G; Korabecny J; Gastaldello S; Lauschke VM
- Article: ADVANCED SCIENCE. 2021;8(16):e2100106Shen JX; Couchet M; Dufau J; de Castro Barbosa T; Ulbrich MH; Helmstadter M; Kemas AM; Zandi Shafagh R; Marques M-A; Hansen JB; Mejhert N; Langin D; Ryden M; Lauschke VM
- Editorial: NEW ENGLAND JOURNAL OF MEDICINE. 2021;385(5):463-465Stebbing J; Lauschke VM
- Article: SCIENCE ADVANCES. 2021;7(1):eabe4724Stebbing J; Nievas GS; Falcone M; Youhanna S; Richardson P; Ottaviani S; Shen JX; Sommerauer C; Tiseo G; Ghiadoni L; Virdis A; Monzani F; Rizos LR; Forfori F; Avendano-Cespedes A; De Marco S; Carrozzi L; Lena F; Sanchez-Jurado PM; Lacerenza LG; Cesira N; Caldevilla-Bernardo D; Perrella A; Niccoli L; Mendez LS; Matarrese D; Goletti D; Tan Y-J; Monteil V; Dranitsaris G; Cantini F; Farcomeni A; Dutta S; Burley SK; Zhang H; Pistello M; Li W; Romero MM; Pretel FA; Simon-Talero RS; Garcia-Molina R; Kutter C; Felce JH; Nizami ZF; Miklosi AG; Penninger JM; Menichetti F; Mirazimi A; Abizanda P; Lauschke VM
- Article: EMBO MOLECULAR MEDICINE. 2020;12(8):e12697Stebbing J; Krishnan V; de Bono S; Ottaviani S; Casalini G; Richardson PJ; Monteil V; Lauschke VM; Mirazimi A; Youhanna S; Tan Y-J; Baldanti F; Sarasini A; Terres JAR; Nickoloff BJ; Higgs RE; Rocha G; Byers NL; Schlichting DE; Nirula A; Cardoso A; Corbellino M
- Article: ADVANCED SCIENCE. 2020;7(15):2000248Oliva-Vilarnau N; Vorrink SU; Ingelman-Sundberg M; Lauschke VM
- Article: SCIENCE TRANSLATIONAL MEDICINE. 2020;12(532):eaaw9709Azzimato V; Jager J; Chen P; Morgantini C; Levi L; Barreby E; Sulen A; Oses C; Willerbrords J; Xu C; Li X; Shen JX; Akbar N; Haag L; Ellis E; Walhen K; Naslund E; Thorell A; Choudhury RP; Lauschke VM; Ryden M; Craige SM; Aouadi M
- Article: CELL. 2018;172(5):1079-1090.e12Sonnen KF; Lauschke VM; Uraji J; Falk HJ; Petersen Y; Funk MC; Beaupeux M; Francois P; Merten CA; Aulehla A
- Article: NATURE. 2013;493(7430):101-105Lauschke VM; Tsiairis CD; Francois P; Aulehla A
All publications from group members
- Article: BIOMATERIALS. 2026;327:123739Nasiri R; Madadelahi M; Nikmaneshi MR; Gokce B; Bijarchi MA; Shah S; Tirpakova Z; Van Gastel D; Taebnia N; de Barros NR; Zhu Y; Morcimen ZG; Gulicli B; Habibey R; Sendemir A; Jain S; Enrico A; Lauschke VM; Dokmeci MR; Pratx G; Khademhosseini A; Herland A
- Journal article: BIOMATERIALS. 2026;327:123739Nasiri R; Madadelahi M; Nikmaneshi MR; Gokce B; Bijarchi MA; Shah S; Tirpáková Z; Van Gastel D; Taebnia N; de Barros NR; Zhu Y; Morcimen ZG; Gulicli B; Habibey R; Sendemir A; Jain S; Enrico A; Lauschke VM; Dokmeci MR; Pratx G; Khademhosseini A; Herland A
- Article: BIOFABRICATION. 2026;18(2)Meyer A; Mayer L-M; Gustafsson L; Eriksson J; Klein K; Büttner FA; Vo DS; Hubert M; Lauschke VM; Sellin ME; Hedhammar M; Artursson P
- Article: CELL COMMUNICATION AND SIGNALING. 2026;24(1):138Lind S; Abdi Ali A; Al Hamoud Al Asswad S; Lauschke VM; Forsman H; Dahlgren C; Johansson LC
- Article: BIOMATERIALS. 2026;331:124089Luo Z; Wu J; Xu B; Wu D; Zhong Y; Hao M; Xu Y; Yang J; Wang Y; Lauschke VM; Ying S; Cheng N
- Journal article: MOLECULAR ASPECTS OF MEDICINE. 2026;107:101430Feng Y; Yang Q; Wang D; Chu Q; Zhou Z; Liu Y; Chen K; Lauschke VM
- Review: MOLECULAR ASPECTS OF MEDICINE. 2026;107:101430Feng Y; Yang Q; Wang D; Chu Q; Zhou Z; Liu Y; Chen K; Lauschke VM
- Journal article: PHARMACOLOGICAL REVIEWS. 2026;:100122Lauschke VM; Ingelman-Sundberg M
- Journal article: BIOMATERIALS. 2026;:124089Luo Z; Wu J; Xu B; Wu D; Zhong Y; Hao M; Xu Y; Yang J; Wang Y; Lauschke VM; Ying S; Cheng N
- Review: ANNUAL REVIEW OF PHARMACOLOGY AND TOXICOLOGY. 2026;66(1):501-518Wright SC; Lindquist P; Rosenkilde MM; Lauschke VM
- Article: ADVANCED SCIENCE. 2026;:e09691Jiang H; Derisoud E; Parreira D; Taebnia N; Jannig PR; Shafagh RZ; Zhao A; Li C; Ortiz M; Maliqueo MA; Stener-Victorin E; Lauschke VM; Deng Q
- Article: NPJ GENOMIC MEDICINE. 2026;11(1):10Krebs K; Luitva LB; Kore AC; Kokasaar R; Joeloo M; Hudjashov G; Maal K; Storset E; Wollmann BM; Karo-Astover L; Fischer K; Team EBR; Lauschke VM; Ingelman-Sundberg M; Molden E; Irs A; Oselin K; Lass J; Milani L
- Article: JOURNAL OF AUTOIMMUNITY. 2026;158:103522Leijding C; Gastaldello S; Schiffer T; Kaewin S; Andreasson KM; Zhong Y; Horuluoglu B; Dastmalchi M; Notarnicola A; Galindo-Feria AS; Lauschke VM; Carlstrom M; Alexanderson H; Lundberg IE; Andersson DC
- Review: ADVANCED BIOLOGY. 2025;9(12):e00337Youhanna S; Taebnia N; Liang Y; Cheng N; Wang Y; Michel M; Lauschke VM
- Article: NAUNYN-SCHMIEDEBERGS ARCHIVES OF PHARMACOLOGY. 2025;:1-16Lyu D; Zhang F; Lauschke VM; Qin J; Gao X; Zhang H; Wang L; Wang G; Wei Y
- Article: ONCOGENE. 2025;44(45):4352-4362De Mattia E; Park Y; Peruzzi E; Zhou Y; Roncato R; Polesel J; Scarabel L; Schwab M; Guchelaar H-J; Swen JJ; Spina M; Puglisi F; Toffoli G; Lauschke VM; Cecchin E
- Article: JOURNAL OF CHEMICAL INFORMATION AND MODELING. 2025;65(21):11950-11964Tzoupis H; Papadourakis M; Papavasileiou KD; Burk O; Lauschke VM; Tsoumanis A; Melagraki G; Afantitis A
- Journal article: STAR PROTOCOLS. 2025;6(4):104183Jian X; Zheng X; Jiao K; Li W; Li X
- Article: ENGINEERING. 2025;54:171-186Sheng H; Liang Y; Lauschke VM; Wang Y
- Article: JOURNAL OF BIOLOGICAL CHEMISTRY. 2025;301(11):110751Gratz L; Turku A; Kozielewicz P; Bowin C-F; Scharf MM; Voss JH; Kinsolving J; Shekhani R; Oliva-Vilarnau N; Koolmeister T; Lauschke VM; Gmeiner P; Schulte G
- Article: PHARMACOLOGICAL RESEARCH. 2025;221:107998Klein K; Burk O; Tremmel R; Buettner FA; Kaestle L; Schroth W; Muerdter TE; Eccles D; Schmidt A-C; Niess H; Zanger UM; Schwab M; Lauschke VM
- Journal article: IMETA. 2025;4(5):e70067Hou W; Hong W; Cai S; Guo D; Yan Z; Zhu J; Shen Y; Wan J; Qu X; Zhang W; Zhao R; Xie Z; Chen Z; Jiang T; Lin Y; Jia W; Wang L; Huang Z; Li X; Tang B
- Article: MOLECULAR AND CELLULAR BIOCHEMISTRY. 2025;480(10):5501-5519Codenotti S; Lauschke VM; Casella EV; Andersson DC; Fanzani A; Gastaldello S
- Doctoral thesis: 2025Kemas A
- Corrigendum: CELL. 2025;188(19):5429-5431Motso A; Pelcman B; Kalinovich A; Kahlous NA; Bokhari MH; Dehvari N; Halleskog C; Waara E; de Jong J; Cheesman E; Kallenberg C; Yakala GK; Murad P; Wetterdal E; Andersson P; van Beek S; Sandstrom A; Alleluia DN; Talamonti E; Youhanna S; Sabatier P; Koenig C; Willems S; Kemas AM; Hutchinson DS; Ham S; Gratz L; Voss J; Marchan-Alvarez JG; Priede M; Jaunsleine K; Spura J; Kovada V; Supe L; Stoddart LA; Holliday ND; Newton PT; Pillon NJ; Schulte G; Summers RJ; Mutule I; Suna E; Olsen JV; Molenaar P; Carlsson J; Lauschke VM; Wright SC; Bengtsson T
- Article: CELL. 2025;188(19):5142-5156.e23Motso A; Pelcman B; Kalinovich A; Kahlous NA; Bokhari MH; Dehvari N; Halleskog C; Waara E; de Jong J; Cheesman E; Kallenberg C; Yakala GK; Murad P; Wetterdal E; Andersson P; van Beek S; Sandström A; Alleluia DN; Talamonti E; Youhanna S; Sabatier P; Koenig C; Willems S; Kemas AM; Hutchinson DS; Ham S; Grätz L; Voss J; Marchan-Alvarez JG; Priede M; Jaunsleine K; Spura J; Kovada V; Supe L; Stoddart LA; Holliday ND; Newton PT; Pillon NJ; Schulte G; Summers RJ; Mutule I; Suna E; Olsen JV; Molenaar P; Carlsson J; Lauschke VM; Wright SC; Bengtsson T
- Review: INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. 2025;26(18):8761Moghazy M; Papathanasiou M; Tzoupis H; Papavasileiou KD; Xing C; Lauschke VM; Afantitis A; Melagraki G
- Journal article: INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. 2025;26(18):8761Moghazy M; Papathanasiou M; Tzoupis H; Papavasileiou KD; Xing C; Lauschke VM; Afantitis A; Melagraki G
- Journal article: HEPATOLOGY. 2025;82(3):702-721Pang Z; Zhang H; Zheng S; Yang X; Liu C; Han Q; Chen Y; Li Z; Zhang X; Cao L; Wang Q; Cao Y; Sun X; Zhao P; Li X; Zheng Q; Sheng R
- Review: ANNALS OF HUMAN GENETICS. 2025;89(5):384-397Zhou Y; Park Y; Camara MD; Lauschke VM
- Article: BIOTECHNOLOGY AND BIOENGINEERING. 2025;122(9):2522-2534Kaellen A; Taebnia N; Widhe M; Lauschke VM; Hedhammar M
- Article: MOLECULAR METABOLISM. 2025;99:102200Smith JAB; Gabriel BM; Brady AJ; Abdelmoez AM; Savikj M; Wright SC; Koutsilieri S; Barres R; Lauschke VM; Krook A; Zierath JR; Pillon NJ
- Preprint: ARXIV. 2025Honoré A; Gálvez BR; Park Y; Zhou Y; Lauschke VM; Xiao M
- Article: BRITISH JOURNAL OF PHARMACOLOGY. 2025;182(14):3353-3370Gasbjerg LS; Rasmussen RS; Dragan A; Lindquist P; Melchiorsen JU; Stepniewski TM; Schiellerup S; Tordrup EK; Gadgaard S; Kizilkaya HS; Willems S; Zhong Y; Wang Y; Wright SC; Lauschke VM; Hartmann B; Holst JJ; Selent J; Rosenkilde MM
- Article: CHEMISTRY-A EUROPEAN JOURNAL. 2025;31(33):e202500382Kehler M; Zhou K; Kemas AM; del Prado A; Hutchinson ES; Nairn EH; Varga M; Plattner Y; Zhong Y; Purewal-Sidhu O; Haslam J; Wiita E; Gildie H; Singerova K; Szaruga Z; Almloef I; Hormann FM; Liu K-C; Wallner O; Ortis F; Homan EJ; Gileadi O; Rudd SG; Stenmark P; de Vega M; Helleday T; D'Arcy-Evans ND; Lauschke VM; Michel M
- Article: CHEMBIOCHEM. 2025;26(11):e202500220Visnes T; Zhou K; Kemas AM; Campopiano D; Lauschke VM; Michel M
- Article: IMETA. 2025;4(3):e70038Pan L; Tang B; Zhang X; Parini P; Tremmel R; Loscalzo J; Lauschke VM; Maron BA; Paci P; Ernberg I; Tan NS; Vegvari A; Liao Z; Rengarajan S; Zubarev R; Fan Y; Zheng X; Jian X; Sheng R; Wang Z; Li X
- Journal article: CLINICAL AND TRANSLATIONAL MEDICINE. 2025;15(5):e70320Peng X; Gao Y; Liu J; Shi X; Li W; Ma Y; Li X; Li H
- Journal article: ENDOCRINE ABSTRACTS. 2025Dekanski A; Li C; Schobloch L; Lindgren E; Wright S; Ferreira D; Lauschke V; Taebnia N; Stener-Victorin E
- Review: EXPERT OPINION ON DRUG METABOLISM & TOXICOLOGY. 2025;21(5):563-577Zhou Y; Zhong Y; Lauschke VM
- Journal article: PHARMACOLOGICAL RESEARCH. 2025;215:107726Huang M; Zhang Y; Chen Z; Yu X; Luo S; Peng X; Li X
- Article: JCI INSIGHT. 2025;10(8):e180943Haag M; Winter S; Kemas AM; Tevini J; Feldman A; Eder SK; Felder TK; Datz C; Paulweber B; Liebisch G; Burk O; Lauschke VM; Aigner E; Schwab M
- Article: CELL REPORTS MEDICINE. 2025;6(4):101992Li X; Pan L; Li W; Liu B; Xiao C; Chew V; Zhang X; Long W; Ginhoux F; Loscalzo J; Buggert M; Zhang X; Sheng R; Wang Z
- Review: PHARMACOGENOMICS. 2025;26(5-6):171-182Tremmel R; Honore A; Park Y; Zhou Y; Xiao M; Lauschke VM
- Journal article: IMETA. 2025;4(3)Comprehensive analysis of multi‐omics single‐cell data using the single‐cell analystPan L; Tang B; Zhang X; Parini P; Tremmel R; Loscalzo J; Lauschke VM; Maron BA; Paci P; Ernberg I; Tan NS; Végvári Á; Liao Z; Rengarajan S; Zubarev R; Fan Y; Zheng X; Jian X; Sheng R; Wang Z; Li X
- Article: DRUG METABOLISM AND DISPOSITION. 2025;53(4):100062Bruecker L; Jacob D; Preiss LC; Zhong Y; Geist F; Hewitt P; Lauschke VM; Petersson C
- Preprint: CHEMRXIV. 2025Kehler M; Zhou K; Kemas A; del Prado A; Scaletti Hutchinson E; Hesslefors Nairn E; Varga M; Plattner Y; Zong Y; Purewal-Sidhu O; Haslam J; Wiita E; Gildie H; Singerova K; Szaruga Z; Almlöf I; Hormann F; Liu K-C; Wallner O; Ortis F; Homan E; Gileadi O; Rudd S; Stenmark P; de Vega M; Helleday T; D'Arcy-Evans N; Lauschke V; Michel M
- Article: MICROORGANISMS. 2025;13(3):534Ova AO; Joffre E; Shafagh RZ; Assuncao MFG; Sidorov RY; Santos LMA; Lauschke VM; Romling U
- Journal article: PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA. 2025;122(7):e2415244122Ma L; Pang Z; Zhang H; Yang X; Zheng S; Chen Y; Ding W; Han Q; Zhang X; Cao L; Fei T; Wang Q; Gao D; He A; Hu K-B; Li X; Sheng R
- Preprint: BIORXIV. 2025Romero AL; Preka E; Pizzirusso G; Arroyo-García LE; Zisiadis GA; Oliva-Vilarnau N; Ruchiy Y; Seitz T; Zhou K; Isla A; Friess L; Sun Y; Canut MQ; Shamikh A; Xu Y; Zhu C; Rodrigues C; Fisahn A; Joseph B; Carlson L-M; Fragkopoulou A; Lauschke V; Betsholtz C; Osman A; Blomgren K
- Review: BRITISH JOURNAL OF CLINICAL PHARMACOLOGY. 2025;91(2):252-263Tremmel R; Pirmann S; Zhou Y; Lauschke VM
- Preprint: BIORXIV. 2025Jiang H; Derisoud E; Parreira D; Taebnia N; Jannig P; Shafagh RZ; Zhao A; Li C; Ortiz M; Maliqueo MA; Stener-Victorin E; Lauschke V; Deng Q
- Preprint: BIORXIV. 2025Codenotti S; Lauschke V; Casella E; Andersson D; Fanzani A; Gastaldello S
- Preprint: BIORXIV. 2025Zhao Q; De Nardo W; Wang R; Zhong Y; Keles U; Sakalauskaite G; Zhao LN; Tay H; Tay H; Youhanna S; Yan M; Xie Y; Kim Y; Lee S; Lim RL; Teo G; Narayanaswamy P; Burton P; Lauschke V; Choi H; Watt M; Kaldis P
- Article: NUCLEIC ACIDS RESEARCH. 2025;53(D1):D1498-D1509Tremmel R; Zhou Y; Camara MD; Laarif S; Eliasson E; Lauschke VM
- Review: DRUG METABOLISM REVIEWS. 2025;57(1):67-90Li K; Lauschke VM; Zhou Y
- Conference publication: PHARMACOGENETICS AND GENOMICS. 2025;35(1):27-28Gonzalez-Padilla D; Zhou Y; Lauschke V
- Article: ADVANCED SCIENCE. 2025;12(3):e2407572Youhanna S; Kemas AM; Wright SC; Zhong Y; Klumpp B; Klein K; Motso A; Michel M; Ziegler N; Shang M; Sabatier P; Kannt A; Sheng H; Oliva-Vilarnau N; Buettner FA; Seashore-Ludlow B; Schreiner J; Windbergs M; Cornillet M; Bjorkstrom NK; Hulsmeier AJ; Hornemann T; Olsen JV; Wang Y; Gramignoli R; Sundstrom M; Lauschke VM
- Book chapter: METHODS IN MOLECULAR BIOLOGY. 2025;p. 205-215Keulen J; Kemas A; Youhanna S; Shafagh RZ; Lauschke VM
- Journal article: COMPUTATIONAL AND STRUCTURAL BIOTECHNOLOGY JOURNAL. 2024;23:3327-3341Zhu B; Li Z; Jin Z; Zhong Y; Lv T; Ge Z; Li H; Wang T; Lin Y; Liu H; Ma T; Wang S; Liao J; Fan X
- Journal article: CHEMICAL ENGINEERING JOURNAL. 2024;499:155987Qiu J; Zhong Y; Shao Y; Zhang G; Yang J; Li Z; Cheng Y
- Article: JOURNAL OF GENETICS AND GENOMICS. 2024;51(11):1228-1236Gonzalez-Padilla D; Camara MD; Lauschke VM; Zhou Y
- Article: ADVANCED SCIENCE. 2024;11(41):e2404510Chen Y; Su Y; Cao X; Siavelis I; Leo IR; Zeng J; Tsagkozis P; Hesla AC; Papakonstantinou A; Liu X; Huang W-K; Zhao B; Haglund C; Ehnman M; Johansson H; Lin Y; Lehtioe J; Zhang Y; Larsson O; Li X; de Flon FH
- Review: PHARMACOLOGICAL REVIEWS. 2024;76(6):1089-1101Weitzberg E; Ingelman-Sundberg M; Lundberg JO; Engberg G; Schulte G; Lauschke VM
- Editorial: SIGNAL TRANSDUCTION AND TARGETED THERAPY. 2024;9(1):269Zhong Y; Lauschke VM
- Journal article: PHYTOCHEMICAL ANALYSIS. 2024;35(7):1649-1658Zhong Y; Wen W; Fan X; Cheng N
- Article: TOXICOLOGY AND APPLIED PHARMACOLOGY. 2024;491:117064He Q; Li M; Ji P; Zheng A; Yao L; Zhu X; Shin J-G; Lauschke VM; Han B; Xiang X
- Article: MOLECULAR THERAPY. 2024;32(9):3012-3024Porebski B; Christ W; Corman A; Haraldsson M; Barz M; Lidemalm L; Haggblad M; Ilmain J; Wright SC; Murga M; Schlegel J; Jarvius M; Lapins M; Sezgin E; Bhabha G; Lauschke VM; Carreras-Puigvert J; Lafarga M; Klingstrom J; Huhn D; Fernandez-Capetillo O
- Journal article: DRUG RESISTANCE UPDATES. 2024;76:101115Hong W-F; Zhang F; Wang N; Bi J-M; Zhang D-W; Wei L-S; Song Z-T; Mills GB; Chen M-M; Li X-X; Du S-S; Yu M
- Journal article: JOURNAL OF CHROMATOGRAPHY A. 2024;1731:465164Zhong Y; Wen W; Luo Z; Cheng N
- Article: CELL DEATH & DISEASE. 2024;15(8):600Guo L; Ma J; Xiao M; Liu J; Hu Z; Xia S; Li N; Yang Y; Gong H; Xi Y; Fu R; Jiang P; Xia C; Lauschke VM; Yan M
- Journal article: NOA. 2024;6(Supplement_1):i37-i38Osman A; Romero AL; Preka E; Pizzirusso G; Arroyo-Garcia LE; Zisiadis GA; Oliva-Vilarnau N; Seitz T; Zhou K; Isla AG; Sun Y; Zhu C; Rodrigues C; Fisahn A; Fragkopoulou A; Lauschke V; Betsholtz C; Blomgren K
- Journal article: ACTA PHARMACEUTICA SINICA B. 2024;14(8):3432-3456Peng X; Fang J; Lou C; Yang L; Shan S; Wang Z; Chen Y; Li H; Li X
- Journal article: NEURO-ONCOLOGY ADVANCES. 2024;6(Suppl 1):i37-i38QSPC-06 TIME-SERIES SINGLE-CELL ANALYSES REVEAL INCESSANT REWIRING OF MICROGLIA AFTER OF CRANIAL IRRADIATION
- Article: NATURE COMPUTATIONAL SCIENCE. 2024;4(8):600-614Yu M; Li W; Yu Y; Zhao Y; Xiao L; Lauschke VM; Cheng Y; Zhang X; Wang Y
- Article: SCIENTIFIC REPORTS. 2024;14(1):17334Mickols E; Meyer A; Handin N; Stuwe M; Eriksson J; Rudfeldt J; Blom K; Fryknas M; Sellin ME; Lauschke VM; Karlgren M; Artursson P
- Article: NATURE COMMUNICATIONS. 2024;15(1):6142Li W; Pan L; Hong W; Ginhoux F; Zhang X; Xiao C; Li X
- Journal article: CELL REPORTS PHYSICAL SCIENCE. 2024;5(7):102093Wu T; Chai N; Chen C; Zhang Z; Wei S; Yang L; Li X; Carvalho RM; Hafeli UO; Peng X; Li H; Gong T
- Article: MOLECULAR METABOLISM. 2024;85:101931Talamonti E; Davegardh J; Kalinovich A; van Beek SMM; Dehvari N; Halleskog C; Bokhari HM; Hutchinson DS; Ham S; Humphrys LJ; Dijon NC; Motso A; Sandstrom A; Zacharewicz E; Mutule I; Suna E; Spura J; Ditrychova K; Stoddart LA; Holliday ND; Wright SC; Lauschke VM; Nielsen S; Scheele C; Cheesman E; Hoeks J; Molenaar P; Summers RJ; Pelcman B; Yakala GK; Bengtsson T
- Article: NATURE METABOLISM. 2024;6(7):1268-1281Kizilkaya HS; Sorensen KV; Madsen JS; Lindquist P; Douros JD; Bork-Jensen J; Berghella A; Gerlach PA; Gasbjerg LS; Mokrosinski J; Mowery SA; Knerr PJ; Finan B; Campbell JE; D'Alessio DA; Perez-Tilve D; Faas F; Mathiasen S; Rungby J; Sorensen HT; Vaag A; Nielsen JS; Holm J-C; Lauenborg J; Damm P; Pedersen O; Linneberg A; Hartmann B; Holst JJ; Hansen T; Wright SC; Lauschke VM; Grarup N; Hauser AS; Rosenkilde MM
- Article: SCIENCE SIGNALING. 2024;17(841):eadi4747Wright SC; Avet C; Gaitonde SA; Muneta-Arrate I; Le Gouill C; Hogue M; Breton B; Koutsilieri S; Alarcia RD; Heroux M; Lauschke VM; Bouvier M
- Article: HEPATOLOGY. 2024;79(6):1337-1351Oliva-Vilarnau N; Beusch CM; Sabatier P; Sakaraki E; Tjaden A; Graetz L; Buettner FA; Dorotea D; Nguyen M; Bergqvist F; Sundstrom Y; Mueller S; Zubarev RA; Schulte G; Tredup C; Gramignoli R; Tietge UJF; Lauschke VM
- Article: NATURE MICROBIOLOGY. 2024;9(5):1499-1512Monteil VM; Wright SC; Dyczynski M; Kellner MJ; Appelberg S; Platzer SW; Ibrahim A; Kwon H; Pittarokoilis I; Mirandola M; Michlits G; Devignot S; Elder E; Abdurahman S; Bereczky S; Bagci B; Youhanna S; Aastrup T; Lauschke VM; Salata C; Elaldi N; Weber F; Monserrat N; Hawman DW; Feldmann H; Horn M; Penninger JM; Mirazimi A
- Article: DRUG METABOLISM AND DISPOSITION. 2024;52(6):539-547Preiss LC; Georgi K; Lauschke VM; Petersson C
- Article: PHARMACOGENOMICS JOURNAL. 2024;24(3):17Zhou Y; Pirmann S; Lauschke VM
- Journal article: DRUG METABOLISM AND DISPOSITION. 2024;52(6):539-547Comparison of Human Long-Term Liver Models for Clearance Prediction of Slowly Metabolized Compounds.Preiss LC; Georgi K; Lauschke VM; Petersson C
- Review: DRUG METABOLISM AND DISPOSITION. 2024;52(6):467-475Ingelman-Sundberg M; Lauschke VM
- Article: ANTIMICROBIAL AGENTS AND CHEMOTHERAPY. 2024;68(5):e01390-e01323Camara MD; Zhou Y; Dara A; Tekete MM; de Sousa TN; Sissoko S; Dembele L; Ouologuem N; Togo AH; Alhousseini ML; Fofana B; Sagara I; Djimde AA; Gil PJ; Lauschke VM
- Article: ADVANCED MATERIALS INTERFACES. 2024;11(14)Harris P; Kaveh J; Kemas AM; Mikaelsson H; Fielden M; Lauschke VM; Shafagh RZ
- Editorial: ARTERIOSCLEROSIS THROMBOSIS AND VASCULAR BIOLOGY. 2024;44(5):1098-1100Taebnia N; Lauschke VM
- Article: ADVANCED SCIENCE. 2024;11(18):e2307734Buniatian GH; Schwinghammer U; Tremmel R; Cynis H; Weiss TS; Weiskirchen R; Lauschke VM; Youhanna S; Ramos I; Valcarcel M; Seferyan T; Rahfeld J-U; Rieckmann V; Klein K; Buadze M; Weber V; Kolak V; Gebhardt R; Friedman SL; Mueller UC; Schwab M; Danielyan L
- Article: EBIOMEDICINE. 2024;103:105124Li X; Hernandez I; Koyuncu S; Haeggblad M; Lidemalm L; Abbas AA; Bendeguz S; Goebloes A; Brautigam L; Lucas JJ; Carreras-Puigvert J; Huehn D; Pircs K; Vilchez D; Fernandez-Capetillo O
- Article: HUMAN GENOMICS. 2024;18(1):40Camara MD; Zhou Y; De Sousa TN; Gil JP; Djimde AA; Lauschke VM
- Article: ISCIENCE. 2024;27(4):109346Di Martino E; Ambikan A; Ramskold D; Umekawa T; Giatrellis S; Vacondio D; Romero AL; Galan MG; Sandberg R; Aden U; Lauschke VM; Neogi U; Blomgren K; Kele J
- Article: GENOME BIOLOGY. 2024;25(1):104Pan L; Parini P; Tremmel R; Loscalzo J; Lauschke V; Maron B; Paci P; Ernberg I; Tan NS; Liao Z; Yin W; Rengarajan S; Li X
- Article: MOLECULAR SYSTEMS BIOLOGY. 2024;20(4):374-402Sommerauer C; Gallardo-Dodd CJ; Savva C; Hases L; Birgersson M; Indukuri R; Shen JX; Carravilla P; Geng K; Sondergaard JN; Ferrer-Aumatell C; Mercier G; Sezgin E; Korach-Andre M; Petersson C; Hagstrom H; Lauschke VM; Archer A; Williams C; Kutter C
- Article: ADVANCED HEALTHCARE MATERIALS. 2024;13(11):2303561Compound Absorption in Polymer Devices Impairs the Translatability of Preclinical Safety AssessmentsKemas AM; Shafagh RZ; Taebnia N; Michel M; Preiss L; Hofmann U; Lauschke VM
- Article: SCIENCE BULLETIN. 2024;69(6):803-822Qian Z-Y; Pan Y-Q; Li X-X; Chen Y-X; Wu H-X; Liu Z-X; Kosar M; Bartek J; Wang Z-X; Xu R-H
- Article: BIOTECHNOLOGY JOURNAL. 2024;19(3):e2300684Koutsilieri S; Mickols E; Vegvari A; Lauschke VM
- Preprint: BIORXIV. 2024Smith JAB; Gabriel B; Abdelmoez A; Savikj M; Wright S; Koutsilieri S; Barrès R; Lauschke V; Krook A; Zierath J; Pillon N
- Review: EXPERT REVIEW OF CLINICAL PHARMACOLOGY. 2024;17(3):213-223Zhou Y; Lauschke VM
- Journal article: MEDICINE PLUS. 2024;1(1):100006Qian Z-Y; Duan C-Y; Cao P-H; Li X-X; Cai Z-Z; Li J-B; Chen P-Y; Xu R-H; Wang Z-X
- Article: BIOTECHNOLOGY JOURNAL. 2024;19(2):e2300587Xing C; Kemas A; Mickols E; Klein K; Artursson P; Lauschke VM
- Preprint: RESEARCH SQUARE. 2024Schulte G; Grätz L; Turku A; Kozielewicz P; Bowin C-F; Scharf M; Voss J; Kinsolving J; Shekhani R; Oliva-Vilarnau N; Koolmeister T; Körber M; Lauschke V; Löber S; Gmeiner P
- Article: NATURE COMMUNICATIONS. 2024;15(1):767Lazzeri-Barcelo F; Oliva-Vilarnau N; Baniol M; Leibiger B; Bergmann O; Lauschke VM; Leibiger IB; Moruzzi N; Berggren P-O
- Review: ANNUAL REVIEW OF PHARMACOLOGY AND TOXICOLOGY. 2024;64:33-51Lauschke VM; Zhou Y; Ingelman-Sundberg M
- Journal article: OPEN RESEARCH EUROPE. 2024;:194Hakomäki H; Pitkänen S; Levonen A-L; Honkakoski P; Greco D; Saarimäki LA; Viegas S; Godinho C; Fyhrquist N; Wincent E; Lauschke V; Hukkanen J; Hakkola J; Vallier L; Fortino V; Afantitis A; Sawatani T; Guzman T; Cnop M; Nawrot T; Harlid S; Vinnars M-T; Tardon A; Grimalt J; Küblbeck J; Rysä J
- Letter: OPEN RESEARCH EUROPE. 2024;4:194Hakomäki H; Pitkänen S; Levonen A-L; Honkakoski P; Greco D; D'Alessandro N; Saarimäki LA; Viegas S; Godinho C; Fyhrquist N; Wincent E; Lauschke VM; Hukkanen J; Hakkola J; Vallier L; Fortino V; Afantitis A; Sawatani T; Guzman TJ; Cnop M; Nawrot T; Harlid S; Vinnars M-T; Tardon A; Grimalt JO; Küblbeck J; Rysä J
- Article: DIABETOLOGIA. 2024;67(1):137-155Ren L; Charbord J; Chu L; Kemas AM; Bertuzzi M; Mi J; Xing C; Lauschke VM; Andersson O
- Review: GUT MICROBES. 2023;15(1):2158034Taebnia N; Romling U; Lauschke VM
- Conference publication: BMJ GLOBAL HEALTH. 2023;8(SUPPL_10):a100.2-a1a101Camara MD; Tekete M; Dama S; de Sousa TN; Kone A; Dinkorma OIT; Bamadio A; Djimde M; Dara A; Fofana B; Ogobara D; Lauschke VM; Borrmann S; Djimde AA; Gil JP
- Article: EMBO JOURNAL. 2023;42(23):e114086Krejcova G; Morgantini C; Zemanova H; Lauschke VM; Kovarova J; Kubasek J; Nedbalova P; Kamps-Hughes N; Moos M; Aouadi M; Dolezal T; Bajgar A
- Article: MOLECULAR BIOLOGY AND EVOLUTION. 2023;40(12):msad267Pan L; Mou T; Huang Y; Hong W; Yu M; Li X
- Editorial: HUMAN GENOMICS. 2023;17(1):105Ingelman-Sundberg M; Nebert DW; Lauschke VM
- Review: CRITICAL REVIEWS IN FOOD SCIENCE AND NUTRITION. 2023;63(28):9436-9481Ghaffari-Bohlouli P; Jafari H; Taebnia N; Abedi A; Amirsadeghi A; Niknezhad SV; Alimoradi H; Jafarzadeh S; Mirzaei M; Nie L; Zhang J; Varma RS; Shavandi A
- Article: ACTA BIOMATERIALIA. 2023;171:336-349Wesseler MF; Taebnia N; Harrison S; Youhanna S; Preiss LC; Kemas AM; Vegvari A; Mokry J; Sullivan GJ; Lauschke VM; Larsen NB
- Editorial: PHARMACOGENOMICS. 2023;24(16):841-844Park Y; Lauschke VM
- Review: JOURNAL OF BIOMATERIALS APPLICATIONS. 2023;38(5):577-604Kafili G; Kabir H; Jalali Kandeloos A; Golafshan E; Ghasemi S; Mashayekhan S; Taebnia N
- Preprint: BIORXIV. 2023Porebski B; Christ W; Corman A; Haraldsson M; Barz M; Lidemalm L; Häggblad M; Ilmain J; Wright S; Murga M; Shlegel J; Sezgin E; Bhabha G; Lauschke V; Lafarga M; Klingström J; Huhn D; Fernandez-Capetillo O
- Article: NATURE COMMUNICATIONS. 2023;14(1):6785Monteil V; Kwon H; John L; Salata C; Jonsson G; Vorrink SU; Appelberg S; Youhanna S; Dyczynski M; Leopoldi A; Leeb N; Volz J; Hagelkruys A; Kellner MJ; Devignot S; Michlits G; Foong-Sobis M; Weber F; Lauschke VM; Horn M; Feldmann H; Elling U; Penninger JM; Mirazimi A
- Article: NATURE COMMUNICATIONS. 2023;14(1):6243Wright SC; Motso A; Koutsilieri S; Beusch CM; Sabatier P; Berghella A; Blondel-Tepaz E; Mangenot K; Pittarokoilis I; Sismanoglou D-C; Le Gouill C; Olsen JV; Zubarev RA; Lambert NA; Hauser AS; Bouvier M; Lauschke VM
- Editorial: HEPATOBILIARY SURGERY AND NUTRITION. 2023;12(5):78084-78784Lauschke VM
- Article: ACS CHEMICAL BIOLOGY. 2023;18(9):1993-2002Novak M; Vajrychova M; Koutsilieri S; Sismanoglou D-C; Kobrlova T; Prchal L; Svobodova B; Korabecny J; Zarybnicky T; Raisova-Stuchlikova L; Skalova L; Lauschke VM; Kucera R; Soukup O
- Article: NPJ GENOMIC MEDICINE. 2023;8(1):24Tremmel R; Zhou Y; Schwab M; Lauschke VM
- Preprint: BIORXIV. 2023Sommerauer C; Gallardo-Dodd C; Savva C; Hases L; Birgersson M; Indukuri R; Shen J; Carravilla P; Geng K; Nørskov Søndergaard J; Ferrer-Aumatell C; Mercier G; Sezgin E; Korach-André M; Petersson C; Hagström H; Lauschke V; Archer A; Williams C; Kutter C
- Journal article: LIVER RESEARCH. 2023;7(3):207-215Shi Y; Wang M; Wu L; Li X; Liao Z
- Conference publication: DIABETOLOGIA. 2023;66(SUPPL 1):S355Yakala GK; Talamonti E; Kalinovich A; van Beek SMM; Holliday ND; Suna E; Mutule I; Wright SC; Lauschke VM; Nielsen S; Scheele C; Summers RJ; Hoeks J; Pelcman B; Bengtsson T
- Conference publication: DIABETOLOGIA. 2023;66(SUPPL 1):S503Moruzzi N; Lazzeri-Barcelo F; Oliva-Vilarnau N; Baniol M; Leibiger B; Bergmann O; Lauschke VM; Leibiger IB; Berggren P-O
- Article: BIOCHEMICAL PHARMACOLOGY. 2023;215:115755Oliva-Vilarnau N; Vorrink SU; Buettner FA; Heinrich T; Sensbach J; Koscielski I; Wienke D; Petersson C; Perrin D; Lauschke VM
- Article: FASEB BIOADVANCES. 2023;5(8):336-353Nilsson G; Mottahedin A; Zelco A; Lauschke VM; Ek CJ; Song J; Ardalan M; Hua S; Zhang X; Mallard C; Hagberg H; Leavenworth JW; Wang X
- Review: CLINICAL AND TRANSLATIONAL MEDICINE. 2023;13(8):e1384Li Y; Wang M; Peng X; Yang Y; Chen Q; Liu J; She Q; Tan J; Lou C; Liao Z; Li X
- Article: NATURE METABOLISM. 2023;5(7):1188-1203Barreby E; Strunz B; Nock S; Naudet L; Shen JX; Johansson H; Soennerborg I; Ma J; Urgard E; Pallett LJ; Hu Y; Fardellas A; Azzimato V; Vankova A; Levi L; Morgantini C; Maini MK; Stal P; Rosshart SP; Coquet JM; Nowak G; Naeslund E; Lauschke VM; Ellis E; Bjoerkstroem NK; Chen P; Aouadi M
- Article: CELL DEATH AND DIFFERENTIATION. 2023;30(7):1666-1678Kanellis DC; Zisi A; Skrott Z; Lemmens B; Espinoza JA; Kosar M; Bjorkman A; Li X; Arampatzis S; Bartkova J; Andujar-Sanchez M; Fernandez-Capetillo O; Mistrik M; Lindstrom MS; Bartek J
- Journal article: SIGNAL TRANSDUCTION AND TARGETED THERAPY. 2023;8(1):234Li X
- Conference publication: JOURNAL OF HEPATOLOGY. 2023;78:S363Lazzeri-Barcelo F; Vilarnau N; Leibiger B; Lauschke V; Leibiger I; Berggren P-O; Moruzzi N
- Conference publication: TISSUE ENGINEERING PART A. 2023;29(11-12):640Wesseler MF; Taebnia N; Larsen NB
- Journal article: JOURNAL OF HEPATOLOGY. 2023;78:s363Lazzeri-Barcelo F; Vilarnau N; Leibiger B; Lauschke V; Leibiger I; Berggren P-O; Moruzzi N
- Article: CURRENT OPINION IN GENETICS & DEVELOPMENT. 2023;80:102057Lauschke VM; Hagberg CE
- Journal article: FRONTIERS IN CELL AND DEVELOPMENTAL BIOLOGY. 2023;11:1209243Mo S-F; Cai Z-Z; Kuai W-H; Li X; Chen Y-T
- Article: PHARMACOGENOMICS JOURNAL. 2023;23(2-3):45-49Skryabin V; Rozochkin I; Zastrozhin M; Lauschke V; Franck J; Bryun E; Sychev D
- Article: STAR PROTOCOLS. 2023;4(2):102260Youhanna S; Bruton J; Jardemark K; Westerblad H; Lauschke VM
- Journal article: SCIENCE BULLETIN. 2023;68(8):763-766Wang Z-X; Pan Y-Q; Li X; Tsubata T; Xu R-H
- Journal article: IUPHAR/BPS GUIDE TO PHARMACOLOGY CITE. 2023;2023(1)Arthofer E; Dijksterhuis J; Grätz L; Hot B; Kozielewicz P; Lauth M; Olofsson J; Petersen J; Polonio T; Schulte G; Strakova K; Valnohova J; Wright S
- Article: ACS CHEMICAL BIOLOGY. 2023;18(4):822-836Tredup C; Ndreshkjana B; Schneider NS; Tjaden A; Kemas AM; Youhanna S; Lauschke VM; Berger B-T; Kraemer A; Berger LM; Roehm S; Knapp S; Farin HF; Mueller S
- Editorial: JOURNAL OF PHYSIOLOGY-LONDON. 2023;601(8):1317-1318Wright SC; Lauschke VM
- Doctoral thesis: 2023Preiss L
- Conference publication: NAUNYN-SCHMIEDEBERGS ARCHIVES OF PHARMACOLOGY. 2023;396:S52Tremmel R; Zhou Y; Nevosadova L; Laarif S; Eliasson E; Lauschke VM
- Conference publication: NAUNYN-SCHMIEDEBERGS ARCHIVES OF PHARMACOLOGY. 2023;396:S52Haag M; Winter S; Kemas AM; Tevini J; Feldman A; Eder S; Felder TK; Datz C; Liebisch G; Burk O; Paulweber B; Lauschke VM; Schwab M; Aigner E
- Conference publication: NAUNYN-SCHMIEDEBERGS ARCHIVES OF PHARMACOLOGY. 2023;396:S52Klein K; Nies AT; Haag M; Hofmann U; Artursson P; Handin N; Burk O; Tremmel R; Winter S; Zhou Y; Schaeffeler E; Zanger UM; Lauschke VM; Schwab M
- Article: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2023;113(3):712-723Khor CC; Winter S; Sutiman N; Muerdter TE; Chen S; Lim JSL; Li Z; Li J; Sim KS; Ganchev B; Eccles D; Eccles B; Tapper W; Zgheib NKK; Tfayli A; Ng RCH; Yap YS; Lim E; Wong M; Wong NS; Ang PCS; Dent R; Tremmel R; Klein K; Schaeffeler E; Zhou Y; Lauschke VMM; Eichelbaum M; Schwab M; Brauch HBB; Chowbay B; Schroth W
- Article: HUMAN GENOMICS. 2023;17(1):15Zhou Y; Nevosadova L; Eliasson E; Lauschke VMM
- Article: JOURNAL OF PHARMACEUTICAL AND BIOMEDICAL ANALYSIS. 2023;224:115154Novak M; Svobodova B; Konecny J; Kuratkova A; Nevosadova L; Prchal L; Korabecny J; Lauschke VM; Soukup O; Kucera R
- Article: LANCET. 2023;401(10374):347-356Swen JJ; van der Wouden CH; Manson LEN; Abdullah-Koolmees H; Blagec K; Blagus T; Boehringer S; Cambon-Thomsen A; Cecchin E; Cheung K-C; Deneer VHM; Dupui M; Ingelman-Sundberg M; Jonsson S; Joefield-Roka C; Just KS; Karlsson MO; Konta L; Koopmann R; Kriek M; Lehr T; Mitropoulou C; Rial-Sebbag E; Rollinson V; Roncato R; Samwald M; Schaeffeler E; Skokou M; Schwab M; Steinberger D; Stingl JC; Tremmel R; Turner RM; van Rhenen MH; Fajardo CLD; Dolzan V; Patrinos GP; Pirmohamed M; Sunder-Plassmann G; Toffoli G; Guchelaar H-J
- Journal article: CURRENT DRUG METABOLISM. 2023;24(2):79Lauschke VM
- Journal article: ZHONGGUO ZHONGYAO ZAZHI. 2023;48(3):829-834Zhong Y; Fan X-H; Li Z-H
- Article: NANOTECHNOLOGY. 2023;34(15):155603Farsani AM; Rahimi F; Taebnia N; Salimi M; Arpanaei A
- Journal article: INTERNATIONAL JOURNAL OF OPHTHALMOLOGY. 2023;16(1):151-155Lou C-Y; Lu L; Li X-X
- Article: NUCLEIC ACIDS RESEARCH. 2023;51(D1):D1019-D1028Pan L; Shan S; Tremmel R; Li W; Liao Z; Shi H; Chen Q; Zhang X; Li X
- Journal article: SPECTROCHIMICA ACTA PART A-MOLECULAR AND BIOMOLECULAR SPECTROSCOPY. 2023;284:121494Ru C; Wen W; Zhong Y
- Article: NORDIC JOURNAL OF PSYCHIATRY. 2023;77(1):73-76Skryabin VY; Franck J; Lauschke VM; Zastrozhin MS; Shipitsyn VV; Bryun EA; Sychev DA
- Article: COMPUTATIONAL AND STRUCTURAL BIOTECHNOLOGY JOURNAL. 2023;21:4361-4369Handin N; Yuan D; Olander M; Wegler C; Karlsson C; Jansson-Lofmark R; Hjelmesaeth J; Asberg A; Lauschke VM; Artursson P
- Conference publication: 2023Lauschke VM
- Article: ALTEX-ALTERNATIVES TO ANIMAL EXPERIMENTATION. 2023;40(3):519-533Turner J; Pound P; Owen C; Hutchinson I; Hop M; Chau DYS; Silva LVB; Coleman M; Dubourg A; Harries LW; Hutter V; Kenna JG; Lauschke VM; Neuhaus W; Roper C; Watkins PB; Welch J; Alvarez LR; Taylor K
- Article: HANDBOOK OF EXPERIMENTAL PHARMACOLOGY. 2023;280:237-260Zhou Y; Lauschke VM
- Article: ISCIENCE. 2022;25(12):105506Xun D; Chen D; Zhou Y; Lauschke VM; Wang R; Wang Y
- Article: ISCIENCE. 2022;25(12):105654Gineste C; Youhanna S; Vorrink SU; Henriksson S; Hernandez A; Cheng AJ; Chaillou T; Buttgereit A; Schneidereit D; Friedrich O; Hultenby K; Bruton JD; Ivarsson N; Sandblad L; Lauschke VM; Westerblad H
- Article: ADVANCED SCIENCE. 2022;9(34):e2203368Shafagh RZ; Youhanna S; Keulen J; Shen JX; Taebnia N; Preiss LC; Klein K; Buettner FA; Bergqvist M; van der Wijngaart W; Lauschke VM
- Article: JOURNAL OF CANCER RESEARCH AND CLINICAL ONCOLOGY. 2022;148(12):3293-3302Xiao Q; Koutsilieri S; Sismanoglou D-C; Lauschke VM
- Article: PHARMACOGENOMICS JOURNAL. 2022;22(5-6):284-293Zhou Y; Lauschke VM
- Review: BASIC & CLINICAL PHARMACOLOGY & TOXICOLOGY. 2022;131(6):452-464Zhou Y; Koutsilieri S; Eliasson E; Lauschke VM
- Review: PHARMACOLOGICAL RESEARCH. 2022;186:106522Xie F; Wang M; Chen Q; Chi T; Zhu S; Wei P; Yang Y; Zhang L; Li X; Liao Z
- Review: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2022;112(6):1159-1171Rodriguez-Antona C; Savieo JL; Lauschke VM; Sangkuhl K; Drogemoller BI; Wang D; Schaik RNH; Gilep AA; Peter AP; Boone EC; Ramey BE; Klein TE; Whirl-Carrillo M; Pratt VM; Gaedigk A
- Editorial: ANNALS OF TRANSLATIONAL MEDICINE. 2022;10(23):1293-0Zhou Y; Lauschke VM
- Article: BIOMEDICINE & PHARMACOTHERAPY. 2022;154:113644De Mattia E; Silvestri M; Polesel J; Ecca F; Mezzalira S; Scarabel L; Zhou Y; Roncato R; Lauschke VM; Calza S; Spina M; Puglisi F; Toffoli G; Cecchin E
- Review: TRENDS IN PHARMACOLOGICAL SCIENCES. 2022;43(10):852-865Zhou Y; Tremmel R; Schaeffeler E; Schwab M; Lauschke VM
- Article: FEBS OPEN BIO. 2022;12(10):1896-1908Colicchia V; Haggblad M; Sirozh O; Porebski B; Balan M; Li X; Lidemalm L; Carreras-Puigvert J; Huhn D; Fernandez-Capetillo O
- Conference publication: 2022;:30-35Sun Y; Zhou Y
- Journal article: REPORTS. 2022;5(3):35Liao Z; Menon D; Zhang L; Lim Y-J; Li W; Li X; Zhao Y
- Journal article: ZHONGGUO ZHONGYAO ZAZHI. 2022;47(17):4545-4550Cheng Y-Y; Wang Y; Liu L; Zhao X-P; Zhong Y; Huang M; Zhang B-L
- Conference publication: TISSUE ENGINEERING PART A. 2022;28:177-178Taebnia N; Zhang R; Andresen TL; Larsen NB
- Article: INTERNATIONAL JOURNAL OF PHARMACEUTICS. 2022;623:121895Erezuma I; Lukin I; Pimenta-Lopes C; Ventura F; Garcia-Garcia P; Reyes R; Arnau MR; Delgado A; Taebnia N; Kadumudi FB; Dolatshahi-Pirouz A; Orive G
- Article: JOURNAL OF PERSONALIZED MEDICINE. 2022;12(7):1187Ivanov HY; Grigorova D; Lauschke VM; Velinov B; Stoychev K; Kyosovska G; Shopov P
- Conference publication: JOURNAL OF HEPATOLOGY. 2022;77:S756-S757Barreby E; Strunz B; Azzimato V; Shen JX; Naudet L; Sonnerborg I; Sulen A; Nock S; Levi L; Morgantini C; Fardellas A; Johansson H; Nowak G; Stal P; Ellis E; Naslund E; Lauschke V; Bjorkstrom N; Chen P; Aouadi M
- Journal article: JOURNAL OF HEPATOLOGY. 2022;77:S665-S666Azzimato V; Chen P; Barreby E; Morgantini C; Levi L; Vankova A; Jager J; Sulen A; Diotallevi M; Shen JX; Ellis E; Ryden M; Naslund E; Thorell A; Lauschke V; Channon KM; Crabtree MJ; Haschemi A; Craige SM; Mori M; Spallotta F; Aouadi M
- Article: AMERICAN JOURNAL OF HUMAN GENETICS. 2022;109(6):1077-1091Trpchevska N; Freidin MB; Broer L; Oosterloo BC; Yao S; Zhou Y; Vona B; Bishop C; Bizaki-Vallaskangas A; Canlon B; Castellana F; Chasman DI; Cherny S; Christensen K; Concas MP; Correa A; Elkon R; Mengel-From J; Gao Y; Giersch ABS; Girotto G; Gudjonsson A; Gudnason V; Heard-Costa NL; Hertzano R; v.B. Hjelmborg J; Hjerling-Leffler J; Hoffman HJ; Kaprio J; Kettunen J; Krebs K; Kahler AK; Lallemend F; Launer LJ; Lee I-M; Leonard H; Li C-M; Lowenheim H; Magnusson PKE; van Meurs J; Milani L; Morton CC; Makitie A; Nalls MA; Nardone GG; Nygaard M; Palviainen T; Pratt S; Quaranta N; Ramo J; Saarentaus E; Sardone R; Satizabal CL; Schweinfurth JM; Seshadri S; Shiroma E; Shulman E; Simonsick E; Spankovich C; Tropitzsch A; Lauschke VM; Sullivan PF; Goedegebure A; Cederroth CR; Williams FMK; Nagtegaal AP
- Article: ADVANCED MATERIALS TECHNOLOGIES. 2022;7(6)Schmidleithner C; Zhang R; Taebnia N; Larsen EKU; Almdal K; Larsen NB
- Review: HUMAN GENETICS. 2022;141(6):1113-1136Zhou Y; Lauschke VM
- Letter: AMERICAN JOURNAL OF HUMAN GENETICS. 2022;109(5):973Wyckelsma VL; Venckunas T; Houweling PJ; Schlittler M; Lauschke VM; Tiong CF; Wood HD; Paulauskas H; Eimantas N; Andersson DC; North KN; Brazaitis M; Westerblad H
- Article: PHARMACEUTICS. 2022;14(5):944Dagli-Hernandez C; Borges JB; Rodrigues Marcal EDS; Costa de Freitas RC; Mori AA; Goncalves RM; Faludi AA; de Oliveira VF; Ferreira GM; Bastos GM; Zhou Y; Lauschke VM; Cerda A; Hirata MH; Crespo Hirata RD
- Review: WORLD JOURNAL OF DIABETES. 2022;13(4):308-318Lin J-R; Wang Z-T; Sun J-J; Yang Y-Y; Li X-X; Wang X-R; Shi Y; Zhu Y-Y; Wang R-T; Wang M-N; Xie F-Y; Wei P; Liao Z-H
- Article: AAPS JOURNAL. 2022;24(2):41Preiss LC; Lauschke VM; Georgi K; Petersson C
- Article: JOURNAL OF THE AMERICAN GERIATRICS SOCIETY. 2022;70(3):650-658Salmeron Rios S; Mas Romero M; Cortes Zamora EB; Tabernero Sahuquillo MT; Romero Rizos L; Manuel Sanchez-Jurado P; Sanchez-Nievas G; Blas Senalada JJ; Garcia Nogueras I; Estrella Cazalla JDD; Andres-Pretel F; Romero AM; Lauschke VM; Stebbing J; Abizanda P; Cespedes AA
- Article: PHARMACOLOGICAL RESEARCH. 2022;176:106087Siamoglou S; Koromina M; Hishinuma E; Yamazaki S; Tsermpini E-E; Kordou Z; Fukunaga K; Chantratita W; Zhou Y; Lauschke VM; Mushiroda T; Hiratsuka M; Patrinos GP
- Review: PHARMACOLOGICAL REPORTS. 2022;74(1):47-66Dagli-Hernandez C; Zhou Y; Lauschke VM; Genvigir FDV; Hirata TDC; Hirata MH; Hirata RDC
- Article: JOURNAL OF NEUROINFLAMMATION. 2022;19(1):20Zhou K; Han J; Lund H; Boggavarapu NR; Lauschke VM; Goto S; Cheng H; Wang Y; Tachi A; Xie C; Zhu K; Sun Y; Osman AM; Liang D; Han W; Gemzell-Danielsson K; Betsholtz C; Zhang X-M; Zhu C; Enge M; Joseph B; Harris RA; Blomgren K
- Journal article: SPECTROCHIMICA ACTA PART A-MOLECULAR AND BIOMOLECULAR SPECTROSCOPY. 2022;264:120250Zhong Y; Ru C; Wang S; Li Z; Cheng Y
- Journal article: EXPERT REVIEW OF PRECISION MEDICINE AND DRUG DEVELOPMENT. 2022;7(1):39-49Kemas AM; Youhanna S; Lauschke VM
- Article: CURRENT DRUG METABOLISM. 2022;23(13):1067-1071Skryabin V; Zastrozhin M; Parkhomenko A; Lauschke VM; Smirnov V; Petukhov A; Pankratenko E; Pozdnyakov S; Koporov S; Denisenko N; Akmalova K; Bryun E; Sychev D
- Book chapter: COMPREHENSIVE PHARMACOLOGY. 2022;p. 53-83Zhou Y; Lauschke VM
- Journal article: CELLULAR AND MOLECULAR GASTROENTEROLOGY AND HEPATOLOGY. 2022;13(5):1592-1609.e9Li H; Qu L; Yang Y; Zhang H; Li X; Zhang X
- Review: PHARMACOLOGICAL REVIEWS. 2022;74(1):141-206Youhanna S; Kemas AM; Preiss L; Zhou Y; Shen JX; Cakal SD; Paqualini FS; Goparaju SK; Shafagh RZ; Lind JU; Sellgren CM; Lauschke VM
- Review: BASIC & CLINICAL PHARMACOLOGY & TOXICOLOGY. 2022;130:5-15Ingelman-Sundberg M; Lauschke VM
- Article: METHODS IN MOLECULAR BIOLOGY. 2022;2445:337-349Kemas AM; Lauschke VM
- Review: ENDOCRINE METABOLIC & IMMUNE DISORDERS-DRUG TARGETS. 2022;22(13):1263-1275Genvigir FDV; Dagli-Hernandez C; Hirata TDC; Zhou Y; Lauschke VM; Hirata MH; Hirata RDC
- Article: GENETICS IN MEDICINE. 2022;24(1):157-169Yang M; Johnsson P; Brautigam L; Yang XR; Thrane K; Gao J; Tobin NP; Zhou Y; Yu R; Nagy N; Engstrom PG; Tuominen R; Eriksson H; Lundeberg J; Tucker MA; Goldstein AM; Egyhazi-Brage S; Zhao J; Cao Y; Hoiom V
- Preprint: BIORXIV. 2021Xun D; Chen D; Zhou Y; Lauschke V; Wang R; Wang Y
- Article: ACS APPLIED MATERIALS & INTERFACES. 2021;13(49):58434-58446Taebnia N; Zhang R; Kromann EB; Dolatshahi-Pirouz A; Andresen TL; Larsen NB
- Letter: CELL RESEARCH. 2021;31(12):1311-1314Xu L; Chen B; Schihada H; Wright SC; Turku A; Wu Y; Han G-W; Kowalski-Jahn M; Kozielewicz P; Bowin C-F; Zhang X; Li C; Bouvier M; Schulte G; Xu F
- Article: JOURNAL OF DRUG DELIVERY SCIENCE AND TECHNOLOGY. 2021;66:102768Shaki H; Vasheghani-Farahani E; Ganji F; Jafarzadeh-Holagh S; Taebnia N; Dolatshahi-Pirouz A
- Article: GASTROENTEROLOGY. 2021;161(6):1982-1997.e11Azzimato V; Chen P; Barreby E; Morgantini C; Levi L; Vankova A; Jager J; Sulen A; Diotallevi M; Shen JX; Miller A; Ellis E; Ryden M; Naslund E; Thorell A; Lauschke VM; Channon KM; Crabtree MJ; Haschemi A; Craige SM; Mori M; Spallotta F; Aouadi M
- Article: PHARMACOLOGICAL RESEARCH. 2021;173:105904Koromina M; Pandi MT; van der Spek PJ; Patrinos GP; Lauschke VM
- Review: MOLECULAR DIAGNOSIS & THERAPY. 2021;25(6):735-755Crespo Hirata TD; Dagli-Hernandez C; Dalla Vecchia Genvigir F; Lauschke VM; Zhou Y; Hirata MH; Crespo Hirata RD
- Conference publication: PEDIATRIC RESEARCH. 2021;90(SUPPL 1):8Hellstrom W; Ekstrom C; Bruschettini M; Carey G; Barton N; Lauschke VM; Vallius-Kvist S; Wang X; Ley D
- Article: JOURNAL OF THE AMERICAN GERIATRICS SOCIETY. 2021;69(10):2752-2758Abizanda P; Calbo Mayo JM; Mas Romero M; Cortes Zamora EB; Tabernero Sahuquillo MT; Romero Rizos L; Sanchez-Jurado PM; Sanchez-Nievas G; Campayo Escolano C; Ochoa Serrano A; Sanchez-Flor Alfaro V; Lopez Bru R; Gomez Ballesteros C; Caldevilla Bernardo D; Callejas Gonzalez FJ; Andres-Pretel F; Lauschke VM; Stebbing J
- Article: SCIENCE ADVANCES. 2021;7(36):eabi6856Zhou Y; Arribas GH; Turku A; Juergenson T; Mkrtchian S; Krebs K; Wang Y; Svobodova B; Milani L; Schulte G; Korabecny J; Gastaldello S; Lauschke VM
- Journal article: IUPHAR/BPS GUIDE TO PHARMACOLOGY CITE. 2021;2021(3)Arthofer E; Dijksterhuis J; Hot B; Kozielewicz P; Lauth M; Olofsson J; Petersen J; Polonio T; Schulte G; Strakova K; Valnohova J; Wright S
- Journal article: ADVANCED MATERIALS. 2021;33(35)Kadumudi FB; Hasany M; Pierchala MK; Jahanshahi M; Taebnia N; Mehrali M; Mitu CF; Shahbazi M; Zsurzsan T; Knott A; Andresen TL; Dolatshahi‐Pirouz A
- Article: ADVANCED MATERIALS. 2021;33(35):e2100047Kadumudi FB; Hasany M; Pierchala MK; Jahanshahi M; Taebnia N; Mehrali M; Mitu CF; Shahbazi M-A; Zsurzsan T-G; Knott A; Andresen TL; Dolatshahi-Pirouz A
- Review: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2021;110(3):626-636Zhou Y; Lauschke VM
- Review: RSC CHEMICAL BIOLOGY. 2021;2(4):1115-1143Larsen JB; Taebnia N; Dolatshahi-Pirouz A; Eriksen AZ; Hjorringgaard C; Kristensen K; Larsen NW; Larsen NB; Marie R; Mundler A-K; Parhamifar L; Urquhart AJ; Weller A; Mortensen KI; Flyvbjerg H; Andresen TL
- Editorial: FRONTIERS IN GENETICS. 2021;12:736626Editorial: Population Pharmacogenomics (PGx): From Variant Identification to Clinical ImplementationHiratsuka M; Zhou Y; Lauschke VM
- Journal article: EUROPEAN JOURNAL OF CLINICAL PHARMACOLOGY. 2021;77(8):1095-1111Wang T; Zhou Y; Cao G
- Article: GENES & DEVELOPMENT. 2021;35(15-16):1190-1207Zelco A; Borjesson V; de Kanter JK; Lebrero-Fernandez C; Lauschke VM; Rocha-Ferreira E; Nilsson G; Nair S; Svedin P; Bemark M; Hagberg H; Mallard C; Holstege FCP; Wang X
- Article: ADVANCED SCIENCE. 2021;8(16):e2100106Shen JX; Couchet M; Dufau J; de Castro Barbosa T; Ulbrich MH; Helmstadter M; Kemas AM; Zandi Shafagh R; Marques M-A; Hansen JB; Mejhert N; Langin D; Ryden M; Lauschke VM
- Article: CURRENT OPINION IN STRUCTURAL BIOLOGY. 2021;69:142-149Wright SC; Bouvier M
- Article: DRUG METABOLISM AND DISPOSITION. 2021;49(8):668-678Preiss LC; Liu R; Hewitt P; Thompson D; Georgi K; Badolo L; Lauschke VM; Petersson C
- Article: CURRENT OPINION IN PHARMACOLOGY. 2021;59:11-18Youhanna S; Wright SC; Lauschke VM
- Editorial: NEW ENGLAND JOURNAL OF MEDICINE. 2021;385(5):463-465Stebbing J; Lauschke VM
- Review: JOURNAL OF CONTROLLED RELEASE. 2021;335:437-448Li X; Li W; Wang M; Liao Z
- Review: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2021;110(1):82-97Desta Z; El-Boraie A; Gong L; Somogyi AA; Lauschke VM; Dandara C; Klein K; Miller NA; Klein TE; Tyndale RF; Whirl-Carrillo M; Gaedigk A
- Conference publication: PROCEEDINGS OF SPIE - THE INTERNATIONAL SOCIETY FOR OPTICAL ENGINEERING. 2021;11786:117861x-117861x-5-65Narag JPC; Taebnia N; Zhang R; Andresen TL; Larsen NB; Kromann EB
- Conference publication: PROCEEDINGS OF SPIE - THE INTERNATIONAL SOCIETY FOR OPTICAL ENGINEERING. 2021;11786:117861r-117861r-5-53Narag JPC; Taebnia N; Zhang R; Andresen TL; Larsen NB; Kromann EB
- Article: ANTIMICROBIAL AGENTS AND CHEMOTHERAPY. 2021;65(7):e0027521-10.1128/aac.00221Pernaute-Lau L; Adegnika AA; Zhou Y; Zinsou JF; Gil JP; Krishna S; Kremsner PG; Lauschke VM; Velavan TP
- Article: NPJ GENOMIC MEDICINE. 2021;6(1):41Xiao Q; Lauschke VM
- Article: INTERNATIONAL JOURNAL OF PHARMACEUTICS. 2021;602:120595Toftdal MS; Taebnia N; Kadumudi FB; Andresen TL; Frogne T; Winkel L; Grunnet LG; Dolatshahi-Pirouz A
- Article: JOURNAL OF THE AMERICAN GERIATRICS SOCIETY. 2021;69(6):1441-1447Salmeron Rios S; Mas Romero M; Cortes Zamora EB; Tabernero Sahuquillo MT; Romero Rizos L; Sanchez-Jurado PM; Sanchez-Nievas G; Senalada JJB; Garcia Nogueras I; Estrella Cazalla JDD; Andres-Pretel F; Murillo Romero A; Lauschke VM; Stebbing J; Abizanda P
- Article: PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA. 2021;118(20):e2025846118Wright SC; Lukasheva V; Le Gouill C; Kobayashi H; Breton B; Mailhot-Larouche S; Blondel-Tepaz E; Vieira NA; Costa-Neto C; Heroux M; Lambert NA; Parreiras-e-Silva LT; Bouvier M
- Article: AMERICAN JOURNAL OF PHYSIOLOGY-CELL PHYSIOLOGY. 2021;320(5):C822-C841Dufau J; Shen JX; Couchet M; Barbosa TDC; Mejhert N; Massier L; Griseti E; Mouisel E; Amri E-Z; Lauschke VM; Ryden M; Langin D
- Article: MAGNESIUM RESEARCH. 2021;34(2):74-83Petrovic J; Labudovic-Borovic M; Vorrink SU; Lauschke VM; Pejuskovic B; Pesic V
- Review: DRUG METABOLISM REVIEWS. 2021;53(2):207-233Yadav J; El Hassani M; Sodhi J; Lauschke VM; Hartman JH; Russell LE
- Review: DRUG METABOLISM REVIEWS. 2021;53(2):253-278Russell LE; Zhou Y; Almousa AA; Sodhi JK; Nwabufo CK; Lauschke VM
- Review: DRUG METABOLISM REVIEWS. 2021;53(2):245-252Lauschke VM
- Preprint: BIORXIV. 2021Gineste C; Youhanna S; Vorrink S; Henriksson S; Hernández A; Cheng A; Chaillou T; Buttgereit A; Schneidereit D; Friedrich O; Hultenby K; Bruton J; Ivarsson N; Sandblad L; Lauschke V; Westerblad H
- Article: AMERICAN JOURNAL OF HUMAN GENETICS. 2021;108(3):446-457Wyckelsma VL; Venckunas T; Houweling PJ; Schlittler M; Lauschke VM; Tiong CF; Wood HD; Ivarsson N; Paulauskas H; Eimantas N; Andersson DC; North KN; Brazaitis M; Westerblad H
- Article: FASEB JOURNAL. 2021;35(3):e21305Kemas AM; Youhanna S; Shafagh RZ; Lauschke VM
- Corrigendum: NATURE METABOLISM. 2021;3(2):287Morgantini C; Jager J; Li X; Levi L; Azzimato V; Sulen A; Barreby E; Xu C; Tencerova M; Naslund E; Kumar C; Verdeguer F; Straniero S; Hultenby K; Bjorkstrom NK; Ellis E; Ryden M; Kutter C; Hurrell T; Lauschke VM; Boucher J; Tomcala A; Krejcova G; Bajgar A; Aouadi M
- Journal article: JOURNAL OF PHARMACEUTICAL AND BIOMEDICAL ANALYSIS. 2021;193:113759Zhong Y; Wang S; Zhu B; Wang R; Cheng Y
- Article: EMBO MOLECULAR MEDICINE. 2021;13(1):e13426Monteil V; Dyczynski M; Lauschke VM; Kwon H; Wirnsberger G; Youhanna S; Zhang H; Slutsky AS; Hurtado del Pozo C; Horn M; Montserrat N; Penninger JM; Mirazimi A
- Journal article: PHARMACOGENOMICS RESEARCH AND PERSONALIZED MEDICINE. 2021;0(0):0Zhou Y; Lauschke VM
- Journal article: JOURNAL OF AUTOIMMUNITY. 2021;116:102571Sheikh AAD; Gomaa S; Li X; Routledge M; Saigoh K; Numoto N; Angata T; Hitomi Y; Takematsu H; Tsuiji M; Ito N; Kusunoki S; Tsubata T
- Review: JOURNAL OF PHARMACEUTICAL SCIENCES. 2021;110(1):50-65Youhanna S; Lauschke VM
- Article: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2021;109(1):160-174Zhou Y; Krebs K; Milani L; Lauschke VM
- Article: SCIENCE ADVANCES. 2021;7(1):eabe4724Stebbing J; Nievas GS; Falcone M; Youhanna S; Richardson P; Ottaviani S; Shen JX; Sommerauer C; Tiseo G; Ghiadoni L; Virdis A; Monzani F; Rizos LR; Forfori F; Avendano-Cespedes A; De Marco S; Carrozzi L; Lena F; Sanchez-Jurado PM; Lacerenza LG; Cesira N; Caldevilla-Bernardo D; Perrella A; Niccoli L; Mendez LS; Matarrese D; Goletti D; Tan Y-J; Monteil V; Dranitsaris G; Cantini F; Farcomeni A; Dutta S; Burley SK; Zhang H; Pistello M; Li W; Romero MM; Pretel FA; Simon-Talero RS; Garcia-Molina R; Kutter C; Felce JH; Nizami ZF; Miklosi AG; Penninger JM; Menichetti F; Mirazimi A; Abizanda P; Lauschke VM
- Article: ACS APPLIED BIO MATERIALS. 2020;3(12):8757-8767Shafagh RZ; Shen JX; Youhanna S; Guo W; Lauschke VM; van der Wijngaart W; Haraldsson T
- Article: BRITISH JOURNAL OF CANCER. 2020;123(12):1782-1789Zhou Y; Dagli Hernandez C; Lauschke VM
- Editorial: JCO ONCOLOGY PRACTICE. 2020;16(12):799-800Koutsilieri S; Patrinos GP
- Editorial: TRENDS IN BIOTECHNOLOGY. 2020;38(12):1312-1315Orive G; Taebnia N; Erezuma I; Andresen TL; Dolatshahi-Pirouz A
- Review: PHARMACOGENOMICS. 2020;21(18):1299-1310Xiao Q; Zhou Y; Lauschke VM
- Article: BIOCHIMICA ET BIOPHYSICA ACTA-MOLECULAR CELL RESEARCH. 2020;1867(12):118849Sundqvist M; Holdfeldt A; Wright SC; Moller TC; Siaw E; Jennbacken K; Franzyk H; Bouvier M; Dahlgren C; Forsman H
- Article: EXPERIMENTAL CELL RESEARCH. 2020;395(2):112234Liu X; Heras G; Lauschke VM; Mi J; Tian G; Gastaldello S
- Preprint: BIORXIV. 2020Wyckelsma VL; Venckunas T; Houweling PJ; Schlittler M; Lauschke VM; Tiong CF; Wood H; Ivarsson N; Paulauskas H; Eimantas N; Andersson DC; North KN; Brazaitis M; Westerblad H
- Article: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2020;108(4):844-855Hendriks DFG; Vorrink SU; Smutny T; Sim SC; Nordling A; Ullah S; Kumondai M; Jones BC; Johansson I; Andersson TB; Lauschke VM; Ingelman-Sundberg M
- Article: PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA. 2020;117(35):21723-21730Okashah N; Wright SC; Kawakami K; Mathiasen S; Zhou J; Lu S; Javitch JA; Inoue A; Bouvier M; Lambert NA
- Editorial: AMERICAN JOURNAL OF MEDICINE. 2020;133(9):e451-e454Parini P; Altucci L; Balligand J-L; Baumbach J; Ferdinandy P; Filetti S; Maron BA; Petrillo E; Silverman EK; Barabasi A-L; Loscalzo J
- Editorial: NPJ SYSTEMS BIOLOGY AND APPLICATIONS. 2020;6(1):29Maron BA; Altucci L; Balligand J-L; Baumbach J; Ferdinandy P; Filetti S; Parini P; Petrillo E; Silverman EK; Barabasi A-L; Loscalzo J
- Article: EMBO MOLECULAR MEDICINE. 2020;12(8):e12697Stebbing J; Krishnan V; de Bono S; Ottaviani S; Casalini G; Richardson PJ; Monteil V; Lauschke VM; Mirazimi A; Youhanna S; Tan Y-J; Baldanti F; Sarasini A; Terres JAR; Nickoloff BJ; Higgs RE; Rocha G; Byers NL; Schlichting DE; Nirula A; Cardoso A; Corbellino M
- Article: ADVANCED SCIENCE. 2020;7(15):2000248Oliva-Vilarnau N; Vorrink SU; Ingelman-Sundberg M; Lauschke VM
- Editorial: PHARMACOLOGICAL RESEARCH. 2020;158:104839Koutsilieri S; Tzioufa F; Sismanoglou D-C; Patrinos GP
- Letter: TRENDS IN PHARMACOLOGICAL SCIENCES. 2020;41(8):503-506Ingelman-Sundberg M; Lauschke VM
- Review: DRUG METABOLISM REVIEWS. 2020;52(3):395-407Russell LE; Schleiff MA; Gonzalez E; Bart AG; Broccatelli F; Hartman JH; Humphreys WG; Lauschke VM; Martin I; Nwabufo C; Prasad B; Scott EE; Segall M; Takahashi R; Taub ME; Sodhi JK
- Article: CELL REPORTS. 2020;31(9):107699Osman AM; Sun Y; Burns TC; He L; Kee N; Oliva-Vilarnau N; Alevyzaki A; Zhou K; Louhivuori L; Uhlen P; Hedlund E; Betsholtz C; Lauschke VM; Kele J; Blomgren K
- Article: HUMAN MUTATION. 2020;41(6):1112-1122Kounelis F; Kanterakis A; Kanavos A; Pandi M-T; Kordou Z; Manusama O; Vonitsanos G; Katsila T; Tsermpini E-E; Lauschke VM; Koromina M; van der Spek PJ; Patrinos GP
- Preprint: BIORXIV. 2020Sundqvist M; Holdfeldt A; Wright S; Møller T; Siaw E; Jennbacken K; Franzyk H; Bouvier M; Dahlgren C; Forsman H
- Article: HUMAN GENETICS. 2020;139(5):623-646Xiao Q; Zhou Y; Lauschke VM
- Article: INTERNATIONAL JOURNAL OF CANCER. 2020;146(9):2475-2487Xiao Q; Zhou Y; Winter S; Buettner F; Schaeffeler E; Schwab M; Lauschke VM
- Preprint: RESEARCH SQUARE. 2020Stebbing J; Krishnan V; de Bono S; Ottaviani S; Casalini G; Richardson P; Monteil V; Lauschke V; Mirazimi A; Terres JR; Nickoloff B; Higgs R; Rocha G; Byers N; Schlichting D; Cardoso A; Corbellino M
- Article: MOLECULAR PHARMACEUTICS. 2020;17(4):1170-1181Russell LE; Zhou Y; Lauschke VM; Kim RB
- Review: NPJ GENOMIC MEDICINE. 2020;5(1):9Lauschke VM; Ingelman-Sundberg M
- Article: PHARMACOLOGICAL RESEARCH. 2020;153:104590Koutsilieri S; Tzioufa F; Sismanoglou D-C; Patrinos GP
- Article: SCIENCE TRANSLATIONAL MEDICINE. 2020;12(532):eaaw9709Azzimato V; Jager J; Chen P; Morgantini C; Levi L; Barreby E; Sulen A; Oses C; Willerbrords J; Xu C; Li X; Shen JX; Akbar N; Haag L; Ellis E; Walhen K; Naslund E; Thorell A; Choudhury RP; Lauschke VM; Ryden M; Craige SM; Aouadi M
- Article: FASEB JOURNAL. 2020;34(2):2269-2286Namuduri AV; Heras G; Lauschke VM; Vitadello M; Traini L; Cacciani N; Gorza L; Gastaldello S
- Review: CHEMICAL RESEARCH IN TOXICOLOGY. 2020;33(1):38-60Shen JX; Youhanna S; Shafagh RZ; Kele J; Lauschke VM
- Review: JOURNAL OF CLINICAL MEDICINE. 2020;9(1):E286-286Xiao Q; Nobre A; Pineiro P; Berciano-Guerrero M-A; Alba E; Cobo M; Lauschke VM; Barragan I
- Article: HUMAN GENOMICS. 2020;14(1):4Koromina M; Koutsilieri S; Patrinos GP
- Letter: BASIC & CLINICAL PHARMACOLOGY & TOXICOLOGY. 2020;126(1):7-8van der Wouden CH; van Rhenen MH; Jama WOM; Ingelman-Sundberg M; Lauschke VM; Konta L; Schwab M; Swen JJ; Guchelaar H-J
- Article: COMPUTATIONAL AND STRUCTURAL BIOTECHNOLOGY JOURNAL. 2020;18:52-58Zhou Y; Lauschke VM
- Article: EUROPEAN JOURNAL OF HUMAN GENETICS. 2020;28(1):88-94Petrovic J; Pesic V; Lauschke VM
- Article: ADVANCED HEALTHCARE MATERIALS. 2020;9(1):e1901023Orive G; Taebnia N; Dolatshahi-Pirouz A
- Journal article: ZHONGGUO ZHONGYAO ZAZHI. 2019;44(24):5269-5276Zhong Y; Ru C-L; Zhang B-L; Cheng Y-Y
- Article: HUMAN GENETICS. 2019;138(11-12):1359-1377Schaller L; Lauschke VM
- Article: PHARMACOGENOMICS. 2019;20(15):1085-1092Lauschke VM; Nordling A; Zhou Y; Fontalva S; Barragan I; Ingelman-Sundberg M
- Article: COLLOIDS AND SURFACES B-BIOINTERFACES. 2019;182:110353Hasany M; Taebnia N; Yaghmaei S; Shahbazi M-A; Mehrali M; Dolatshahi-Pirouz A; Arpanaei A
- Article: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2019;106(4):866-873van der Wouden CH; van Rhenen MH; Jama WOM; Ingelman-Sundberg M; Lauschke VM; Konta L; Schwab M; Swen JJ; Guchelaar H-J
- Review: FRONTIERS IN PHARMACOLOGY. 2019;10:1093Zhou Y; Shen JX; Lauschke VM
- Journal article: IUPHAR/BPS GUIDE TO PHARMACOLOGY CITE. 2019;2019(4)Arthofer E; Dijksterhuis J; Hot B; Kozielewicz PL; Lauth M; Olofsson J; Petersen J; Polonio T; Schulte G; Strakova K; Valnohova J; Wright S
- Review: ADVANCED SCIENCE. 2019;6(16):1801664Talebian S; Mehrali M; Taebnia N; Pennisi CP; Kadumudi FB; Foroughi J; Hasany M; Nikkhah M; Akbari M; Orive G; Dolatshahi-Pirouz A
- Review: EXPERT OPINION ON BIOLOGICAL THERAPY. 2019;19(8):773-779Carmen Echave M; Hernaez-Moya R; Iturriaga L; Luis Pedraz J; Lakshminarayanan R; Dolatshahi-Pirouz A; Taebnia N; Orive G
- Article: FRONTIERS IN PHARMACOLOGY. 2019;10:830Simeonidis S; Koutsilieri S; Vozikis A; Cooper DN; Mitropoulou C; Patrinos GP
- Journal article: ADVANCED SCIENCE. 2019;6(16):1970094Talebian S; Mehrali M; Taebnia N; Pennisi CP; Kadumudi FB; Foroughi J; Hasany M; Nikkhah M; Akbari M; Orive G; Dolatshahi‐Pirouz A
- Conference publication: EUROPEAN JOURNAL OF HUMAN GENETICS. 2019;27:545-546Santos M; Niemi M; Hiratsuka M; Kumondai M; Ingelman-Sundberg M; Lauschke VM; Rodriguez-Antona C
- Article: ARTERIOSCLEROSIS THROMBOSIS AND VASCULAR BIOLOGY. 2019;39(7):1432-1447Jensen LD; Hot B; Ramskold D; Germano RFV; Yokota C; Giatrellis S; Lauschke VM; Hubmacher D; Li MX; Hupe M; Arnold TD; Sandberg R; Frisen J; Trusohamn M; Martowicz A; Wisniewska-Kruk J; Nyqvist D; Adams RH; Apte SS; Vanhollebeke B; Stenman JM; Kele J
- Review: BIOTECHNOLOGY JOURNAL. 2019;14(7):e1800347Lauschke VM; Shafagh RZ; Hendriks DFG; Ingelman-Sundberg M
- Editorial: AMERICAN JOURNAL OF HEMATOLOGY. 2019;94(7):737-740Koutsilieri S; Caudle KE; Alzghari SK; Monte AA; Relling MV; Patrinos GP
- Article: MICROSYSTEMS & NANOENGINEERING. 2019;5:25Shafagh RZ; Decrop D; Ven K; Vanderbeke A; Hanusa R; Breukers J; Pardon G; Haraldsson T; Lammertyn J; van der Wijngaart W
- Article: GENETICS IN MEDICINE. 2019;21(6):1345-1354Reisberg S; Krebs K; Lepamets M; Kals M; Magi R; Metsalu K; Lauschke VM; Vilo J; Milani L
- Review: PHARMACOLOGY & THERAPEUTICS. 2019;197:122-152Lauschke VM; Zhou Y; Ingelman-Sundberg M
- Corrigendum: NATURE METABOLISM. 2019;1(4):497Morgantini C; Jager J; Li X; Levi L; Azzimato V; Sulen A; Barreby E; Xu C; Tencerova M; Naslund E; Kumar C; Verdeguer F; Straniero S; Hultenby K; Bjorkstrom NK; Ellis E; Ryden M; Kutter C; Hurrell T; Lauschke VM; Boucher J; Tomcala A; Krejcova G; Bajgar A; Aouadi M
- Article: NATURE METABOLISM. 2019;1(4):445-459Morgantini C; Jager J; Li X; Levi L; Azzimato V; Sulen A; Barreby E; Xu C; Tencerova M; Naslund E; Kumar C; Verdeguer F; Straniero S; Hultenby K; Bjorkstrom NK; Ellis E; Ryden M; Kutter C; Hurrell T; Lauschke VM; Boucher J; Tomcala A; Krejcova G; Bajgar A; Aouadi M
- Article: PHARMACOGENOMICS JOURNAL. 2019;19(2):115-126Zhou Y; Mkrtchian S; Kumondai M; Hiratsuka M; Lauschke VM
- Article: EUROPEAN JOURNAL OF PHARMACEUTICAL SCIENCES. 2019;130:65-77Lauschke VM; Ingelman-Sundberg M
- Article: ADVANCED SCIENCE. 2019;6(5):1801241Kadumudi FB; Jahanshahi M; Mehrali M; Zsurzsan T-G; Taebnia N; Hasany M; Mohanty S; Knott A; Godau B; Akbari M; Dolatshahi-Pirouz A
- Journal article: ADVANCED SCIENCE. 2019;6(5):1970026Kadumudi FB; Jahanshahi M; Mehrali M; Zsurzsan T; Taebnia N; Hasany M; Mohanty S; Knott A; Godau B; Akbari M; Dolatshahi‐Pirouz A
- Article: EUROPEAN JOURNAL OF HUMAN GENETICS. 2019;27(3):442-454Tasa T; Krebs K; Kals M; Magi R; Lauschke VM; Haller T; Puurand T; Remm M; Esko T; Metspalu A; Vilo J; Milani L
- Article: NATURE COMMUNICATIONS. 2019;10(1):667Wright SC; Kozielewicz P; Kowalski-Jahn M; Petersen J; Bowin C-F; Slodkowicz G; Marti-Solano M; Rodriguez D; Hot B; Okashah N; Strakova K; Valnohova J; Babu MM; Lambert NA; Carlsson J; Schulte G
- Article: PHARMACOLOGICAL RESEARCH. 2019;139:550-559Zhang B; Lauschke VM
- Article: DATABASE-THE JOURNAL OF BIOLOGICAL DATABASES AND CURATION. 2019;2019:baz085Brown P; Tan A-C; El-Esawi MA; Liehr T; Blanck O; Gladue DP; Almeida GMF; Cernava T; Sorzano CO; Yeung AWK; Engel MS; Chandrasekaran AR; Muth T; Staege MS; Daulatabad SV; Widera D; Zhang J; Meule A; Honjo K; Pourret O; Yin C-C; Zhang Z; Cascella M; Flegel WA; Goodyear CS; van Raaij MJ; Bukowy-Bieryllo Z; Campana LG; Kurniawan NA; Lalaouna D; Huttner FJ; Ammerman BA; Ehret F; Cobine PA; Tan E-C; Han H; Xia W; McCrum C; Dings RPM; Marinello F; Nilsson H; Nixon B; Voskarides K; Yang L; Costa VD; Bengtsson-Palme J; Bradshaw W; Grimm DG; Kumar N; Martis E; Prieto D; Sabnis SC; Amer SEDR; Liew AWC; Perco P; Rahimi F; Riva G; Zhang C; Devkota HP; Ogami K; Basharat Z; Fierz W; Siebers R; Tan K-H; Boehme KA; Brenneisen P; Brown JAL; Dalrymple BP; Harvey DJ; Ng G; Werten S; Bleackley M; Dai Z; Dhariwal R; Gelfer Y; Hartmann MD; Miotla P; Tamaian R; Govender P; Gurney-Champion OJ; Kauppila JH; Zhang X; Echeverria N; Subhash S; Sallmon H; Tofani M; Bae T; Bosch O; Cuiv PO; Danchin A; Diouf B; Eerola T; Evangelou E; Filipp FV; Klump H; Kurgan L; Smith SS; Terrier O; Tuttle N; Ascher DB; Janga SC; Schulte LN; Becker D; Browngardt C; Bush SJ; Gaullier G; Ide K; Meseko C; Werner GDA; Zaucha J; Al-Farha AA; Greenwald NF; Popoola SI; Rahman MS; Xu J; Yang SY; Hiroi N; Alper OM; Baker CI; Bitzer M; Chacko G; Debrabant B; Dixon R; Forano E; Gilliham M; Kelly S; Klempnauer K-H; Lidbury BA; Lin MZ; Lynch I; Ma W; Maibach EW; Mather DE; Nandakumar KS; Ohgami RS; Parchi P; Tressoldi P; Xue Y; Armitage C; Barraud P; Chatzitheochari S; Coelho LP; Diao J; Doxey AC; Gobet A; Hu P; Kaiser S; Mitchell KM; Salama MF; Shabalin IG; Song H; Stevanovic D; Yadollahpour A; Zeng E; Zinke K; Alimba CG; Beyene TJ; Cao Z; Chan SS; Gatchell M; Kleppe A; Piotrowski M; Torga G; Woldesemayat AA; Cosacak MI; Haston S; Ross SA; Williams R; Wong A; Abramowitz MK; Effiong A; Lee S; Abid MB; Agarabi C; Alaux C; Albrecht DR; Atkins GJ; Beck CR; Bonvin AMJJ; Bourke E; Brand T; Braun RJ; Bull JA; Cardoso P; Carter D; Delahay RM; Ducommun B; Duijf PHG; Epp T; Eskelinen E-L; Fallah M; Farber DB; Fernandez-Triana J; Feyerabend F; Florio T; Friebe M; Furuta S; Gabrielsen M; Gruber J; Grybos M; Han Q; Heinrich M; Helantera H; Huber M; Jeltsch A; Jiang F; Josse C; Jurman G; Kamiya H; de Keersmaecker K; Kristiansson E; de Leeuw F-E; Li J; Liang S; Lopez-Escamez JA; Lopez-Ruiz FJ; Marchbank KJ; Marschalek R; Martin CS; Miele AE; Montagutelli X; Morcillo E; Nicoletti R; Niehof M; O'Toole R; Ohtomo T; Oster H; Palma J-A; Paterson R; Peifer M; Portilla M; Portillo MC; Pritchard AL; Pusch S; Raghava GPS; Roberts NJ; Ross K; Schuele B; Sergeant K; Shen J; Stella A; Sukocheva O; Uversky VN; Vanneste S; Villet MH; Viveiros M; Vorholt JA; Weinstock C; Yamato M; Zabetakis I; Zhao X; Ziegler A; Aizat WM; Atlas L; Bridges KM; Chakraborty S; Deschodt M; Domingues HS; Esfahlani SS; Falk S; Guisado JL; Kane NC; Kueberuwa G; Lau CL; Liang D; Liu E; Luu AM; Ma C; Ma L; Moyer R; Norris AD; Panthee S; Parsons JR; Peng Y; Pinto IM; Reschke CR; Sillanpaa E; Stewart CJ; Uhle F; Yang H; Zhou K; Zhu S; Ashry M; Bergsland N; Berthold M; Chen C-E; Colella V; Cuypers M; Eskew EA; Fan X; Gajda M; Gonzalezlez-Prendes R; Goodin A; Graham EB; Groen EJN; Gutierrez-Sacristan A; Habes M; Heffler E; Higginbottom DB; Janzen T; Jayaraman J; Jibb LA; Jongen S; Kinyanjui T; Koleva-Kolarova RG; Li Z; Liu Y-P; Lund BA; Lussier AA; Ma L; Mier P; Moore MD; Nagler K; Orme MW; Pearson JA; Prajapati AS; Saito Y; Troder SE; Uchendu F; Verloh N; Voutchkova DD; Abu-Zaid A; Bakkach J; Baumert P; Dono M; Hanson J; Herbelet S; Hobbs E; Kulkarni A; Kumar N; Liu S; Loft ND; Reddan T; Senghore T; Vindin H; Xu H; Bannon R; Chen B; Cheung JTK; Cooper J; Esnakul AK; Feghali KA; Ghelardi E; Gnasso A; Horbar J; Lai HM; Li J; Ma L; Ma R; Pan Z; Peres MA; Pranata R; Seow E; Sydes M; Testoni I; Westermair AL; Yang Y; Afnan M; Albiol J; Albuquerque LG; Amiya E; Amorim RM; An Q; Andersen SU; Aplin JD; Argyropoulos C; Asmann YW; Assaeed AM; Atanasov AG; Atchison DA; Avery SV; Avillach P; Baade PD; Backman L; Badie C; Baldi A; Ball E; Bardot O; Barnett AG; Basner M; Batra J; Bazanova OM; Beale A; Beddoe T; Bell ML; Berezikov E; Berners-Price S; Bernhardt P; Berry E; Bessa TB; Billington C; Birch J; Blakely RD; Blaskovich MAT; Blum R; Boelaert M; Bogdanos D; Bosch C; Bourgoin T; Bouvard D; Boykin LM; Bradley G; Braun D; Brownlie J; Bruhl A; Burt A; Butler LM; Byrareddy SN; Byrne HJ; Cabantous S; Calatayud S; Candal E; Carlson K; Casillas S; Castelvetro V; Caswell PT; Cavalli G; Cerovsky V; Chagoyen M; Chen C-S; Chen DF; Chen H; Chen H; Chen J-T; Chen Y; Cheng C; Cheng J; Chinapaw M; Chinopoulos C; Cho WCS; Chong L; Chowdhury D; Chwalibog A; Ciresi A; Cockcroft S; Conesa A; Cook PA; Cooper DN; Coqueret O; Corea EM; Costa E; Coupland C; Crawford SY; Cruz AD; Cui H; Cui Q; Culver DC; D'Angiulli A; Dahms TES; Daigle F; Dalgleish R; Danielsen HE; Darras S; Davidson SM; Day DA; Degirmenci V; Demaison L; Devriendt K; Ding J; Dogan Y; Dong XC; Donner CF; Dressick W; Drevon CA; Duan H; Ducho C; Dumaz N; Dwarakanath BS; Ebell MH; Eisenhardt S; Elkum N; Engel N; Erickson TB; Fairhead M; Faville MJ; Fejzo MS; Festa F; Feteira A; Flood-Page P; Forsayeth J; Fox SA; Franks SJ; Frentiu FD; Frilander MJ; Fu X; Fujita S; Galea I; Galluzzi L; Gani F; Ganpule AP; Garcia-Alix A; Gedye K; Giordano M; Giunta C; Gleeson PA; Goarant C; Gong H; Gora D; Gough MJ; Goyal R; Graham KE; Grande-Perez A; Graves PM; Greidanus H; Grice D; Grunau C; Gumulya Y; Guo Y; Gurevich VV; Gusev O; Hacker E; Hage SR; Hagen G; Hahn S; Haller DM; Hammerschmidt S; Han J; Han R; Handfield M; Hapuarachchi HC; Harder T; Hardingham JE; Heck M; Heers M; Hew KF; Higuchi Y; St Hilaire C; Hilton R; Hodzic E; Hone A; Hongoh Y; Hu G; Huber HP; Hueso LE; Huirne J; Hurt L; Idborg H; Ikeo K; Ingley E; Jakeman PM; Jensen A; Jia H; Jia H; Jia S; Jiang J; Jiang X; Jin Y; Jo D; Johnson AM; Johnston M; Jonscher KR; Jorens PG; Jorgensen JOL; Joubert JW; Jung S-H; Junior AM; Kahan T; Kamboj SK; Kang Y-K; Karamanos Y; Karp NA; Kelly R; Kenna R; Kennedy J; Kersten B; Khalaf RA; Khalid JM; Khatlani T; Khider T; Kijanka GS; King SRB; Kluz T; Knox P; Kobayashi T; Koch K-W; Kohonen-Corish MRJ; Kong X; Konkle-Parker D; Korpela KM; Kostrikis LG; Kraiczy P; Kratz H; Krause G; Krebsbach PH; Kristensen SR; Kumari P; Kunimatsu A; Kurdak H; Kwon YD; Lachat C; Lagisz M; Laky B; Lammerding J; Lange M; Larrosa M; Laslett AL; LeClair EE; Lee K-W; Lee M-Y; Lee M-S; Li G; Li J; Lieb K; Lim YY; Lindsey ML; Line P-D; Liu D; Liu F; Liu H; Liu H; Lloyd VK; Lo T-W; Locci E; Loidl J; Lorenzen J; Lorkowski S; Lovell NH; Lu H; Lu W; Lu Z; Luengo GS; Lundh L-G; Lysy PA; Mabb A; Mack HG; Mackey DA; Mahdavi SR; Maher P; Maher T; Maity SN; Malgrange B; Mamoulakis C; Mangoni AA; Manke T; Manstead ASR; Mantalaris A; Marsal J; Marschall H-U; Martin FL; Martinez-Raga J; Martinez-Salas E; Mathieu D; Matsui Y; Maza E; McCutcheon JE; Mckay GJ; McMillan B; McMillan N; Meads C; Medina L; Merrick BA; Metzger DW; Meunier FA; Michaelis M; Micheau O; Mihara H; Mintz EM; Mizukami T; Moalic Y; Mohapatra DP; Monteiro A; Montes M; Moran JV; Morozov SY; Mort M; Murai N; Murphy DJ; Murphy SK; Murray SA; Naganawa S; Nammi S; Nasios G; Natoli RM; Nguyen F; Nicol C; van Nieuwerburgh F; Nilsen EB; Nobile CJ; O'Mahony M; Ohlsson S; Olatunbosun O; Olofsson P; Ortiz A; Ostrikov K; Otto S; Outeiro TF; Ouyang S; Paganoni S; Page A; Palm C; Paradies Y; Parsons MH; Parsons N; Pascal P; Paul E; Peckham M; Pedemonte N; Pellizzon MA; Petrelli M; Pichugin A; Pinto CJC; Plevris JN; Pollesello P; Polz M; Ponti G; Porcelli P; Prince M; Quinn GP; Quinn TJ; Ramula S; Rappsilber J; Rehfeldt F; Reiling JH; Remacle C; Rezaei M; Riddick EW; Ritter U; Roach NW; Roberts DD; Robles G; Rodrigues T; Rodriguez C; Roislien J; Roobol MJ; Rowe JA; Ruepp A; van Ruitenbeek J; Rust P; Saad S; Sack GH; Santos M; Saudemont A; Sava G; Schrading S; Schramm A; Schreiber M; Schuler S; Schymkowitz J; Sczyrba A; Seib KL; Shi H-P; Shimada T; Shin J-S; Shortt C; Silveyra P; Skinner D; Small I; Smeets PAM; So P-W; Solano F; Sonenshine DE; Song J; Southall T; Speakman JR; Srinivasan MV; Stabile LP; Stasiak A; Steadman KJ; Stein N; Stephens AW; Stewart DI; Stine K; Storlazzi C; Stoynova NV; Strzalka W; Suarez OM; Sultana T; Sumant AV; Summers MJ; Sun G; Tacon P; Tanaka K; Tang H; Tanino Y; Targett-Adams P; Tayebi M; Tayyem R; Tebbe CC; Telfer EE; Tempel W; Teodorczyk-Injeyan JA; Thijs G; Thorne S; Thrift AG; Tiffon C; Tinnefeld P; Tjahjono DH; Tolle F; Toth E; del Tredici AL; Tsapas A; Tsirigotis K; Turak A; Tzotzos G; Udo EE; Utsumi T; Vaidyanathan S; Vaillant M; Valsesia A; Vandenbroucke RE; Veiga FH; Vendrell M; Vesk PA; Vickers P; Victor VM; Villemur R; Vohl M-C; Voolstra CR; Vuillemin A; Wakelin S; Waldron L; Walsh LJ; Wang AY; Wang F; Wang Y; Watanabe Y; Weigert A; Wen J-C; Wham C; White EP; Wiener J; Wilharm G; Wilkinson S; Willmann R; Wilson C; Wirth B; Wojan TR; Wolff M; Wong BM; Wu T-W; Wuerbel H; Xiao X; Xu D; Xu JW; Xu J; Xue B; Yalcin S; Yan H; Yang E-C; Yang S; Yang W; Ye Y; Ye Z-Q; Yli-Kauhaluoma J; Yoneyama H; Yu Y; Yuan G-C; Yuh C-H; Zaccolo M; Zeng C; Zevnik B; Zhang C; Zhang L; Zhang L; Zhang Y; Zhang Y; Zhang Z; Zhang Z-Y; Zhao Y; Zhou M; Zuberbier T; Aanei CM; Ahmad R; Al-Lawama M; Alanio A; Allardyce J; Alonso-Caneiro D; Atack JM; Baier D; Bansal A; Benezeth Y; Berbesque C; Berrevoet F; Biedermann PHW; Bijleveld E; Bittner F; Blombach F; Van den Bos W; Boudreau SA; Bramoweth AD; Braubach O; Cai Y; Campbell M; Cao Z; Catry T; Chen X; Cheng S; Chung H-J; Chavez-Fumagalli MA; Conway A; Costa BM; Cyr N; Dean LT; Denzel MS; Dlamini SV; Dudley KJ; Dufies M; Ecke T; Eckweiler D; Eixarch E; El-Adawy H; Emmrich JV; Eustace AJ; Falter-Wagner CM; Fuss J; Gao J; Gill MR; Gloyn L; Goggs R; Govinden U; Greene G; Greiff V; Grundle DS; Gruneberg P; Gumede N; Haore G; Harrison P; Hoenner X; Hojsgaard D; Hori H; Ikonomopoulou MP; Jeurissen P; Johnson DM; Kabra D; Kamagata K; Karmakar C; Kasian O; Kaye LK; Khan MM; Kim Y-M; Kish JK; Kobold S; Kohanbash G; Kohls G; Kugler J-M; Kumar G; Lacy-Colson J; Latif A; Lauschke VM; Li B; Lim CJ; Liu F; Liu X; Lu J-J; Lu Q; Mahavadi P; Marzocchi U; McGarrigle CA; van Meerten T; Min R; Moal I; Molari M; Molleman L; Mondal SR; Van de Mortel T; Moss WN; Moultos OA; Mukherjee M; Nakayama K; Narayan E; Navaratnarajah; Neumann P-A; Nie J; Nie Y; Niemeyer F; Nolan F; Nwaiwu O; Oldenmenger WH; Olumayede E; Ou J; Pallebage-Gamarallage M; Pearce SP; Pelkonen T; Pelleri MC; Pereira JL; Pheko M; Pinto KA; Piovesan A; Pluess M; Podolsky IM; Prescott J; Qi D; Qi X; Raikou VD; Ranft A; Rhodes J; Rotge J-Y; Rowe AD; Saggar M; Schuon RA; Shahid S; Shalchyan V; Shirvalkar P; Shiryayev O; Singh J; Smout MJ; Soares A; Song C; Srivastava K; Srivastava RK; Sun J; Szabo A; Szymanski W; Tai CNP; Takeuchi H; Tanadini-Lang S; Tang F; Tao W; Theron G; Tian CF; Tian Y-S; Tuttle LM; Valenti A; Verlot P; Walker M; Wang J; Welter D; Winslade M; Wu D; Wu Y-R; Xiao H; Xu B; Xu J; Xu Z; Yang D; Yang M; Yankilevich P; You Y; Yu C; Zhan J; Zhang G; Zhang K; Zhang T; Zhang Y; Zhao G; Zhao J; Zhou X; Zhu Z; Ajani PA; Anazodo UC; Bagloee SA; Bail K; Bar I; Bathelt J; Benkeser D; Bernier ML; Blanchard AM; Boakye DW; Bonatsos V; Boon MH; Bouboulis G; Bromfield E; Brown J; Bul KCM; Burton KJ; Butkowski EG; Carroll G; Chao F; Charrier EE; Chen X; Chen Y-C; Chenguang; Choi JR; Christoffersen T; Comel JC; Cosse C; Cui Y; van Dessel P; Dhaval; Diodato D; Duffey M; Dutt A; Egea LG; El-Said M; Faye M; Fernandez-Fernandez B; Foley KG; Founou LL; Fu F; Gadelkareem RA; Galimov E; Garip G; Gemmill A; Gouil Q; Grey J; Gridneva Z; Grothe MJ; Grebert T; Guerrero F; Guignard L; Haenssgen MJ; Hasler D; Holgate JY; Huang A; Hulse-Kemp AM; Jean-Quartier C; Jeon S-M; Jia Y; Jutzeler C; Kalatzis P; Karim M; Karsay K; Keitel A; Kempe A; Keown JR; Khoo CM; Khwaja N; Kievit RA; Kosanic A; Koutoukidis DA; Kramer P; Kumar D; Kirag N; Lanza G; Le TD; Leem JW; Leightley D; Leite A; Lercher L; Li Y; Lim R; Lima LRA; Lin L; Ling T; Liu Y; Liu Z; Lu Y; Lum FM; Luo H; Machhi J; Macleod A; Macwan I; Madala HR; Madani N; de Maio N; Makowiecki K; Mallinson DJ; Margelyte R; Maria C; Markonis Y; Marsili L; Mavoa S; McWilliams L; Megersa M; Mendes CSM; Menichetti J; Mercieca-Bebber R; Miller JJ; Minde D-PM; Minges A; Mishra E; Mishra VR; Moores C; Morrice N; Moskalensky AE; Navarin N; Negera E; Nolet P; Nordberg A; Norden R; Nowicki JP; Olova N; Olszewski P; Onzima R; Pan C-L; Park C; Park DI; Park S; Patil CD; Pedro SA; Perry SR; Peter J; Peterson BM; Pezzuolo A; Pozdnyakov I; Qian S; Qin L; Rafe A; Raote I; Raza A; Rebl H; Refai O; Regan T; Richa T; Richardson MF; Robinson KR; Rossoni L; Rouet R; Safaei S; Schneeberger PHH; Schwotzer D; Sebastian A; Selinski J; Seltmann S; Sha F; Shalev N; Shang J-L; Singer J; Singh M; Smith T; Solomon-Moore E; Song L; Soraggi S; Stanley R; Steckhan N; Strobl F; Subissi L; Supriyanto I; Surve CR; Suzuki T; Syme C; Sorelius K; Tang Y; Tantawy M; Tennakoon S; Teseo S; Toelzer C; Tomov N; Tovar M; Tran L; Tripathi S; Tuladhar AM; Ukubuiwe AC; Ung COL; Valgepea K; Vatanparast H; Vidal A; Wang F; Wang Q; Watari R; Webster R; Webster R; Wei J; Wibowo D; Wingenbach TSH; Xavier RM; Xiao S; Xiong P; Xu S; Xu S; Yao R; Yao W; Yin Q; Yu Y; Zaitsu M; Zeineb Z; Zhan X-Y; Zhang J; Zhang R; Zhang W; Zhang X; Zheng S; Zhou B; Zhou X; Ahmad H; Akinwumi SA; Albery GF; Alhowimel A; Ali J; Alshehri M; Alsuhaibani M; Anikin A; Azubuike SO; Bach-Mortensen A; Baltiansky L; Bartas M; Belachew KY; Bhardwaj V; Binder K; Bland NS; Boah M; Bullen B; Calabro GE; Callahan TJ; Cao B; Chalmers K; Chang W; Che Z; Chen ATY; Chen H; Chen H; Chen Y; Chen Z; Choi Y; Chowdhury MAK; Christensen MR; Cooke RSC; Cottini M; Covington NV; Cunningham C; Delarocque J; Devos L; Dhar AR; Ding K-F; Dong K; Dong Z; Dreyer N; Ekstrand C; Fardet T; Feleke BE; Feurer T; Freitas A; Gao T; Asefa NG; Giganti F; Grabowski P; Guerra-Mora JR; Guo C; Guo X; Gupta H; He S; Heijne M; Heinemann S; Hogrebe A; Huang Z; Iskander-Rizk S; Iyer LM; Jahan Y; James AS; Joel E; Joffroy B; Jegousse C; Kambondo G; Karnati P; Kaya C; Ke A; Kelly D; Kickert R; Kidibule PE; Kieselmann JP; Kim HJ; Kitazawa T; Lamberts A; Li Y; Liang H; Linn SN; Litfin T; Liusuo W; Lygirou V; Mahato AK; Mai Z-M; Major RW; Mali S; Mallis P; Mao W; Mao W; Marvin-Dowle K; Marvin-Dowle K; Mason LD; Merideth B; Merino-Plaza MJ; Merlaen B; Messina R; Mishra AK; Muhammad J; Musinguzi C; Nanou A; Naqash A; Nguyen JT; Nguyen TTH; Ni D; Nida; Notcovich S; Ohst B; Ollivier QR; Osses DF; Peng X; Plantinga A; Pulia M; Rafiq M; Raman A; Raucher-Chene D; Rawski R; Ray A; Razak LA; Rudolf K; Rusch P; Sadoine ML; Schmidt A; Schurr R; Searles S; Sharma S; Sheehan B; Shi C; Shohayeb B; Sommerlad A; Strehlow J; Sun X; Sundar R; Taherzadeh G; Tahir NDM; Tang J; Testa J; Tian Z; Tingting Q; Verheijen GP; Vickstrom C; Wang T; Wang X; Wang Z; Wei P; Wilson A; Wyart; Yassine A-A; Yousefzadeh A; Zare A; Zeng Z; Zhang C; Zhang H; Zhang L; Zhang T; Zhang W; Zhang Z; Zhou J; Zhu D; Adamo V; Adeyemo AA; Aggelidou M; Al-Owaifeer AM; Al-Riyami AZ; Alzghari SK; Andersen V; Angus K; Asaduzzaman M; Asady H; Ato D; Bai X; Baines RL; Ballantyne M; Ban B; Beck J; Ben-Nafa W; Black E; Blancher A; Blankstein R; Bodagh N; Borges PAV; Brooks A; Brox-Ponce J; Brunetti A; Canham CD; Carninci P; Carvajal R; Chang SC; Chao J; Chatterjee P; Chen H; Chen Y-C; Chhatriwalla AK; Chikowe I; Chuang T-J; Collevatti RG; Valera-Cornejo DA; Cuenda A; Dao M; Dauga D; Deng Z; Devkota K; Doan LV; Elewa YHA; Fan D; Faruk M; Feifei S; Ferguson TS; Fleres F; Foster EJ; Foster CS; Furer T; Gao Y; Garcia-Rivera EJ; Gazdar A; George RB; Ghosh S; Gianchecchi E; Gleason JM; Hackshaw A; Hall A; Hall R; Harper P; Hogg WE; Huang G; Hunter KE; IJzerman AP; Jesus C; Jian G; Lewis JSJ; Kanj SS; Kaur H; Kelly S; Kheir F; Kichatova VS; Kiyani M; Klein R; Kovesi T; Kraschnewski JL; Kumar AP; Labutin D; Lazo-Langner A; Leclercq G; Li M; Li Q; Li T; Li Y; Liao W-T; Liao Z-Y; Lin J; Lizer J; Lobreglio G; Lowies C; Lu C; Majeed H; Martin A; Martinez-Sobrido L; Meresh E; Middelveen M; Mohebbi A; Mota J; Mozaheb Z; Muyaya L; Nandhakumar A; Ng SHX; Obeidat M; Oh D-H; Owais M; Pace-Asciak P; Panwar A; Park C; Patterson C; Penagos-Tabaree F; Pianosi PT; Pinzi V; Pridans C; Psaroulaki A; Pujala RK; Pulido-Arjona L; Qi P-F; Rahman P; Rai NK; Rassaf T; Refardt J; Ricciardi W; Riess O; Rovas A; Sacks FM; Saleh S; Sampson C; Schmutz A; Sepanski R; Sharma N; Singh M; Spearman P; Subramaniapillai M; Swali R; Tan CM; Tellechea JI; Thomas L-M; Tong X; Veys R; Vitriol V; Wang H-D; Wang J; Wang J; Waugh J; Webb SA; Williams BA; Workman AD; Xiang T; Xie L-X; Xu J; Xu T; Yang C; Yoon JG; Yuan CM; Zaritsky A; Zhang Y; Zhao H; Zuckerman H; Lyu R; Pullan W; Zhou Y
- Article: METHODS IN MOLECULAR BIOLOGY. 2019;1940:77-95Sibbritt T; Osteil P; Fan X; Sun J; Salehin N; Knowles H; Shen J; Tam PPL
- Review: FRONTIERS IN PHARMACOLOGY. 2018;9:1437Zhou Y; Fujikura K; Mkrtchian S; Lauschke VM
- Article: SCIENCE SIGNALING. 2018;11(559):eaar5536Wright SC; Canizal MCA; Benkel T; Simon K; Le Gouill C; Matricon P; Namkung Y; Lukasheva V; Koenig GM; Laporte SA; Carlsson J; Kostenis E; Bouvier M; Schulte G; Hoffmann C
- Article: ACS NANO. 2018;12(10):9940-9946Shafagh RZ; Vastesson A; Guo W; van der Wijngaart W; Haraldsson T
- Article: ACS APPLIED MATERIALS & INTERFACES. 2018;10(41):34924-34941Hasany M; Thakur A; Taebnia N; Kadumudi FB; Shahbazi M-A; Pierchala MK; Mohanty S; Orive G; Andresen TL; Foldager CB; Yaghmaei S; Arpanaei A; Gaharwar AK; Mehrali M; Dolatshahi-Pirouz A
- Article: JOURNAL OF LIPID RESEARCH. 2018;59(10):1987-2000Zhou Y; Magi R; Milani L; Lauschke VM
- Article: SCIENTIFIC REPORTS. 2018;8(1):14297Kozyra M; Johansson I; Nordling A; Ullah S; Lauschke VM; Ingelman-Sundberg M
- Letter: TOXICOLOGICAL SCIENCES. 2018;165(1):4-5Ingelman-Sundberg M; Lauschke VM
- Article: GENESIS. 2018;56(9):e23246Sibbritt T; Ip CK; Khoo P-L; Wilkie E; Jones V; Sun JQJ; Shen JX; Peng G; Han J-DJ; Jing N; Osteil P; Ramialison M; Tam PPL; Fossat N
- Review: TRENDS IN PHARMACOLOGICAL SCIENCES. 2018;39(9):828-842Schulte G; Wright SC
- Article: PHARMACOGENOMICS. 2018;19(14):1133-1138Ingelman-Sundberg M; Lauschke VM
- Article: JOURNAL OF MEDICAL GENETICS. 2018;55(9):617-627Zhou Y; Lauschke VM
- Conference publication: BASIC & CLINICAL PHARMACOLOGY & TOXICOLOGY. 2018;123:89Santos M; Niemi M; Hiratsuka M; Kumondai M; Ingelman-Sundberg M; Lauschke VM; Rodriguez-Antona C
- Article: PHARMACOGENOMICS. 2018;19(12):931-936Jukic MM; Lauschke VM; Saito T; Hiratsuka M; Ingelman-Sundberg M
- Preprint: BIORXIV. 2018Reisberg S; Krebs K; Kals M; Mägi R; Metsalu K; Lauschke V; Vilo J; Milani L
- Review: FRONTIERS IN MEDICINE. 2018;5:192Oliva-Vilarnau N; Hankeova S; Vorrink SU; Mkrtchian S; Andersson ER; Lauschke VM
- Article: TOXICOLOGICAL SCIENCES. 2018;163(2):655-665Vorrink SU; Zhou Y; Ingelman-Sundberg M; Lauschke VM
- Article: GENETICS IN MEDICINE. 2018;20(6):622-629Santos M; Niemi M; Hiratsuka M; Kumondai M; Ingelman-Sundberg M; Lauschke VM; Rodriguez-Antona C
- Article: HUMAN GENOMICS. 2018;12(1):26Ingelman-Sundberg M; Mkrtchian S; Zhou Y; Lauschke VM
- Journal article: PHYTOMEDICINE. 2018;44:129-137Zhong Y; Zhu J; Yang Z; Shao Q; Fan X; Cheng Y
- Conference publication: BASIC & CLINICAL PHARMACOLOGY & TOXICOLOGY. 2018;123:5Zhou Y; Mkrtchian S; Kumondai M; Hiratsuka M; Lauschke V
- Article: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2018;103(5):745-748Lauschke VM; Ingelman-Sundberg M
- Article: TOXICOLOGICAL SCIENCES. 2018;162(2):655-666Bell CC; Dankers ACA; Lauschke VM; Sison-Young R; Jenkins R; Rowe C; Goldring CE; Park K; Regan SL; Walker T; Schofield C; Baze A; Foster AJ; Williams DP; van de Ven AWM; Jacobs F; van Houdt J; Lahteenmaki T; Snoeys J; Juhila S; Richert L; Ingelman-Sundberg M
- Article: APPLIED IN VITRO TOXICOLOGY. 2018;4(1):1-12Messner S; Fredriksson L; Lauschke VM; Roessger K; Escher C; Bober M; Kelm JM; Ingelman-Sundberg M; Moritz W
- Article: CELL. 2018;172(5):1079-1090.e12Sonnen KF; Lauschke VM; Uraji J; Falk HJ; Petersen Y; Funk MC; Beaupeux M; Francois P; Merten CA; Aulehla A
- Review: ANNUAL REVIEW OF PHARMACOLOGY AND TOXICOLOGY. 2018;58:161-185Lauschke VM; Barragan I; Ingelman-Sundberg M
- Review: ANNUAL REVIEW OF PHARMACOLOGY AND TOXICOLOGY. 2018;58:65-82Ewart L; Dehne E-M; Fabre K; Gibbs S; Hickman J; Hornberg E; Ingelman-Sundberg M; Jang K-J; Jones DR; Lauschke VM; Marx U; Mettetal JT; Pointon A; Williams D; Zimmermann W-H; Newham P
- Journal article: PROCEEDINGS FOR ANNUAL MEETING OF THE JAPANESE PHARMACOLOGICAL SOCIETY. 2018;WCP2018(0):po4-9-16Vorrink SU; Zhou Y; Ingelman-Sundberg M; Lauschke VM
- Article: JOURNAL OF MICROMECHANICS AND MICROENGINEERING. 2017;27(12):124001Sandstrom N; Shafagh RZ; Gylfason KB; Haraldsson T; van der Wijngaart W
- Review: AAPS JOURNAL. 2017;20(1):4Lauschke VM; Milani L; Ingelman-Sundberg M
- Article: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2017;102(4):688-700Zhou Y; Ingelman-Sundberg M; Lauschke VM
- Article: ACS APPLIED MATERIALS & INTERFACES. 2017;9(36):30267-30272Marinins A; Shafagh RZ; van der Wijngaart W; Haraldsson T; Linnros J; Veinot JGC; Popov S; Sychugov I
- Review: TRENDS IN PHARMACOLOGICAL SCIENCES. 2017;38(9):765-770Lauschke VM; Ivanov M; Ingelman-Sundberg M
- Article: NATURE COMMUNICATIONS. 2017;8(1):226Petersen J; Wright SC; Rodriguez D; Matricon P; Lahav N; Vromen A; Friedler A; Stromqvist J; Wennmalm S; Carlsson J; Schulte G
- Editorial: CLINICAL SCIENCE. 2017;131(15):2059-2062Lauschke VM
- Article: OPTICS LETTERS. 2017;42(11):2157-2160Lobov GS; Marinins A; Shafagh RZ; Zhao Y; van der Wijngaart W; Wosinski L; Thylen L; Toprak MS; Haraldsson T; Ostling M; Popov S
- Article: FASEB JOURNAL. 2017;31(6):2696-2708Vorrink SU; Ullah S; Schmidt S; Nandania J; Velagapudi V; Beck O; Ingelman-Sundberg M; Lauschke VM
- Article: DRUG METABOLISM AND DISPOSITION. 2017;45(4):419-429Bell CC; Lauschke VM; Vorrink SU; Palmgren H; Duffin R; Andersson TB; Ingelman-Sundberg M
- Article: ACS APPLIED MATERIALS & INTERFACES. 2017;9(12):10418-10426Decrop D; Pardon G; Brancato L; Kil D; Shafagh RZ; Kokalj T; Haraldsson T; Puers R; van der Wijngaart W; Lammertyn J
- Article: ARCHIVES OF TOXICOLOGY. 2017;91(3):1385-1400Sison-Young RL; Lauschke VM; Johann E; Alexandre E; Antherieu S; Aerts H; Gerets HHJ; Labbe G; Hoet D; Dorau M; Schofield CA; Lovatt CA; Holder JC; Stahl SH; Richert L; Kitteringham NR; Jones RP; Elmasry M; Weaver RJ; Hewitt PG; Ingelman-Sundberg M; Goldring CE; Park BK
- Article: BIOCHEMICAL AND BIOPHYSICAL RESEARCH COMMUNICATIONS. 2017;482(3):399-407Lauschke VM; Mkrtchian S; Ingelman-Sundberg M
- Article: GENETICS IN MEDICINE. 2017;19(1):20-29Kozyra M; Ingelman-Sundberg M; Lauschke VM
- Article: LANGMUIR. 2016;32(50):13394-13402Taebnia N; Morshedi D; Yaghmaei S; Aliakbari F; Rahimi F; Arpanaei A
- Review: CHEMICAL RESEARCH IN TOXICOLOGY. 2016;29(12):1936-1955Lauschke VM; Hendriks DFG; Bell CC; Andersson TB; Ingelman-Sundberg M
- Article: PLOS ONE. 2016;11(12):e0166330Rajabi M; Roxhed N; Shafagh RZ; Haraldson T; Fischer AC; van der Wijngaart W; Stemme G; Niklaus F
- Article: HEPATOLOGY. 2016;64(5):1743-1756Lauschke VM; Vorrink SU; Moro SML; Rezayee F; Nordling A; Hendriks DFG; Bell CC; Sison-Young R; Park BK; Goldring CE; Ellis E; Johansson I; Mkrtchian S; Andersson TB; Ingelman-Sundberg M
- Review: INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. 2016;17(10):E1714-1714Lauschke VM; Ingelman-Sundberg M
- Article: NUCLEIC ACIDS RESEARCH. 2016;44(14):6756-6769Ivanov M; Kals M; Lauschke V; Barragan I; Ewels P; Kaller M; Axelsson T; Lehtio J; Milani L; Ingelman-Sundberg M
- Article: PHARMACOGENOMICS. 2016;17(8):917-924Lauschke VM; Ingelman-Sundberg M
- Article: SCIENTIFIC REPORTS. 2016;6:25187Bell CC; Hendriks DFG; Moro SML; Ellis E; Walsh J; Renblom A; Puigvert LF; Dankers ACA; Jacobs F; Snoeys J; Sison-Young RL; Jenkins RE; Nordling A; Mkrtchian S; Park BK; Kitteringham NR; Goldring CEP; Lauschke VM; Ingelman-Sundberg M
- Article: CLINICAL PHARMACOLOGY & THERAPEUTICS. 2016;99(2):172-185Kalman LV; Agundez JAG; Appell ML; Black JL; Bell GC; Boukouvala S; Bruckner C; Bruford E; Caudle K; Coulthard SA; Daly AK; Del Tredici AL; den Dunnen JT; Drozda K; Everts RE; Flockhart D; Freimuth RR; Gaedigk A; Hachad H; Hartshorne T; Ingelman-Sundberg M; Klein TE; Lauschke VM; Maglott DR; McLeod HL; McMillin GA; Meyer UA; Mueller DJ; Nickerson DA; Oetting WS; Pacanowski M; Pratt VM; Relling MV; Roberts A; Rubinstein WS; Sangkuhl K; Schwab M; Scott SA; Sim SC; Thirumaran RK; Toji LH; Tyndale RF; van Schaik RHN; Whirl-Carrillo M; Yeo KTJ; Zanger UM
- Editorial: TRENDS IN PHARMACOLOGICAL SCIENCES. 2016;37(2):85-86Lauschke VM; Ingelman-Sundberg M
- Journal article: MOLECULAR BIOSYSTEMS. 2015;12(2):606-613Wang C-H; Zhong Y; Zhang Y; Liu J-P; Wang Y-F; Jia W-N; Wang G-C; Li Z; Zhu Y; Gao X-M
- Article: PHARMACOGENETICS AND GENOMICS. 2015;25(12):584-594Fujikura K; Ingelman-Sundberg M; Lauschke VM
- Article: JOURNAL OF MICROMECHANICS AND MICROENGINEERING. 2015;25(7):075002Sandstrom N; Shafagh RZ; Vastesson A; Carlborg CF; van der Wijngaart W; Haraldsson T
- Article: RSC ADVANCES. 2015;5(75):60966-60974Taebnia N; Morshedi D; Doostkam M; Yaghmaei S; Aliakbari F; Singh G; Arpanaei A
- Article: SCIENTIFIC REPORTS. 2014;4:6978Zhou Y; Zhou P; Xin Y; Wang J; Zhu Z; Hu J; Wei S; Ma H
- Journal article: ZHONGGUO ZHONGYAO ZAZHI. 2014;39(17):3287-3290Zhong Y; Zhu J-Q; Fan X-H; Kang L-Y; Li Z
- Journal article: ZHONGGUO ZHONGYAO ZAZHI. 2014;39(14):2660-2664Zhu J-Q; Pan W-F; Zhong Y; Fan X-H; Kang L-Y; Li Z
- Journal article: ZHONGGUO ZHONGYAO ZAZHI. 2014;39(13):2495-2497Zhong Y; Zhu J-Q; Fan X-H; Kang L-Y; Li Z
- Article: ACS APPLIED MATERIALS & INTERFACES. 2014;6(11):8313-8319Molecular Composition, Grafting Density and Film Area Affect the Swelling-Induced Au-S Bond BreakageLv B; Zhou Y; Cha W; Wu Y; Hu J; Li L; Chi L; Ma H
- Article: OPTICS EXPRESS. 2013;21(18):21293-21298Errando-Herranz C; Saharil F; Mola Romero A; Sandstrom N; Shafagh RZ; van der Wijngaart W; Haraldsson T; Gylfason KB
- Article: NATURE. 2013;493(7430):101-105Lauschke VM; Tsiairis CD; Francois P; Aulehla A
- Journal article: CHEST. 2012;142(4):1080a-1080cNeophytou A; Anagnostopoulos N; Vardavas C; Koutsilieri S; Dotis G; Evangelopoulou V; Laden F; Behrakis P; Dockery D
- Article: ANALYST. 2011;136(8):1690-1696Chen X; Zu Y; Xie H; Kemas AM; Gao Z
- Show more
Funding
- Swedish Research Council
- Innovative Medicines Initiative-2 (IMI2) Project EUbOPEN
- Knut and Alice Wallenberg Foundation
- Organ-On-A-Chip Technologies Network Grant
- Horizon 2020 program of the European Union (U-PGx)
In addition, the group acknowledges generous support from the Robert Bosch Foundation for Medical Research (RBMF), Karolinska Institutet, Eli Lilly and Company and Merck KGaA.
Staff and contact
Group leader
- Volker LauschkeProfessor
All members of the group
- Tountzai ElmaliAffiliated to Research
- Domnica-Gabriela GurauPhd Student
- Sofiia IskrakPhd Student
- Javad KavehResearch Assistant
- Aurino KemasAffiliated to Research
- Jibbe KeulenPhd Student
- Stefania KoutsilieriPhd Student
- Xuexin LiAffiliated to Research
- Yingxin LiangAffiliated to Research
- Aikaterini MotsoAffiliated to Research
- Yoomi ParkAffiliated to Research
- Maximilian RathAffiliated to Research
- Elisabeth RohdePhd Student
- Jinhye RyuPhd Student
- Nayere TaebniaResearch Specialist
- Sabine WillemsPostdoctoral Researcher
- Shane WrightAssistant Professor
- Chen XingPhd Student
- Sonia YouhannaAffiliated to Research
- Reza Zandi ShafaghResearch Specialist
- Yi ZhongPostdoctoral Studies
- Yitian ZhouPostdoctoral Researcher
Pharmacoepigenetics

Our ultimate goal is to understand the pharmacological response and identify new biomarkers that are amenable for targeted manipulation. Our strategy is to combine genetic and epigenetic state-of-the-art analyses of clinical samples and functional studies in advanced three-dimensional cell cultures.
We developed a method that distinguishes DNA methylation from hydroxymethylation, and identified variants specific to each of these cytosine modifications important for drug metabolism (Nucleic Acids Research, 2016).
We have recently applied this principle to the identification of the first epigenetic marker of response to cancer immunotherapy (Lancer Resp. Med., 2018, Best Practice, 2019). In addition, we have identified a novel transcriptomic signature of B cell functionality associated with survival in melanoma patients treated with immunotherapy (IJMS 2023).
Another interesting aspect of our research is Abscopal Response, a distant response in a non-irradiated tumor after radiotherapy in other lesions. We have reported a 65% Abscopal Response rate that was associated with an improved Progression-Free Survival (21.2 months vs 3 months, p < 0.0001) in patients that were treated with the combination of Radiotherapy and Immunotherapy (International Journal of Radiation Oncology, 2022).
Our recent projects incorporate this combination therapy with the aim of characterizing the immunogenic cell death that Immune Checkpoint Blockade triggers in those patients developing Abscopal Response. In a preliminary cohort, we have correlated a high percentage of CD4+ and CD8+PDL1 and a low percentage of CD8+PD1+ with a good response. Longitudinally, we also found that an increase in circulating free DNA (cfDNA) concentration during SABR related to a poor response, whereas an increase in CD20+ cells and a decrease in CD19+ cells after the first SABR fraction were associated with a favorable response. Furthermore, we have identified a set of microRNAs (miRNAs) in patients with non-small cell lung cancer that are predictive of response (Zafra J et al., 2024).
Specialised methods
Chicas-sett R et al. Cancers (Basel). 2020;12(8):2178–98.
- Three-dimensional cultures (and treatment/phenotyping) of primary human liver cells and cancer cell lines.
- High-throughput 3D cultures in 96-well plates using hanging drop technology (© InSphero) or Ultra Low Attachment surfaces.
- TET-assisted bisulfite conversion (TAB)-based determination of DNA methylation and hydroxymethylation:
- TAB-EPIC Illumina DNA methylation arrays for genome-wide analysis of 850K CpG sites
- TAB-Methyl-Seq Agilent targeted libraries for Next Generation Sequencing
- TAB-Sanger sequencing
- Adaptations of the protocol for FFPE and cell-free DNA
- TET-independent restriction-based determination of DNA methylation and hydroxymethylation
- Quantification of global DNA methylation and hydroxymethylation by mass spectrometry/liquid chromatography.
- Smart-seq library preparation for single cell RNA-sequencing.
- All RNA-sequencing by ribosomal depletion
- Exome sequencing with DNA from FFPE tumors and paired PBMCs
- ChIP-qPCR for histone modifications in 3D liver cancer cell lines and primary hepatocyte microtissues.
- Bioinformatics for genome-wide and customised transcriptomic/epigenomic analyses. OMICS integration. AI-based prediction models.
- Deciphering the EPIgenetic Signatures of the ABscopal Response towards precision immunotherapy (EPI-SABR, PRYES247250BARR). PI: M Isabel Barragan Mallofret. (IBIMA-Plataforma BIONAND). Spanish Association Against Cancer (AECC). 01/12/2024-30/11/2027. 150.000 €.
- EPIPREDICT: Development of a panel of epigenetic markers in liquid biopsy for prediction of response and prognosis in the context of novel immunotherapy approaches. (DTS23/00114). Instituto de Salud Carlos III. PI: M Isabel Barragán Mallofret (IBIMA-Plataforma BIONAND). 01/01/2024- 31/12/2026. 238.480 €.
- MICROMICS: Ex vivo characterization of the mechanisms associated with Abscopal Response (GEM 2023). PI: M Isabel Barragán Mallofret (IBIMA-Plataforma BIONAND). Melanoma Spanish Group. 01/01/2024-31/12/2025. 40.000 €.
- Multi-modal and Multi-scale Modeling based on Artificial Intelligence Pathways and Spatial Transcriptomics to reveal Lung Cancer Distant Relapse predictors (MAPS-LUNG) (CNS2023-145629). PI: M Isabel Barragán Mallofret (IBIMA-Plataforma BIONAND). Ministry of Science, Innovation and Universities. 01/01/2024-31/12/2025. 399.605 €.
- STINT China-Sweden: Identification of DNA methylation and hydroxymethylation variation and molecular network in primary central nervous system lymphoma
- PI22/01816: Immunotherapy resistance biomarker: integrating Abscopal Response and epigenetics in a non-invasive precision medicine strategy. Health Institute Carlos III.
- ProyExcel_01002: Precision pharmacogenomics of immunotherapy treatment of lung and head and neck cancer. Andalusian Government, Science.
- PI22/01816: Immunotherapy resistance biomarker: integrating Abscopal Response and epigenetics in a non-invasive precision medicine strategy. Health Institute Carlos III.
- SEOM21: MicroRNA signatures in the early detection of the field cancerization in squamous cell oral cavity carcinoma.
- AECC 2020: Radio-immunotherapy in metastatic cancer.
- P20-01326: ORIGEN: Early detection of liver metastases in breast cancer based on the study of methylation and hydroximetylation patterns in the circulating free DNA in blood. Andalusian Government, Science.
- PI18/01592: Omics integration for precision cancer immunotherapy. Health Institute Carlos III.
- Metabolic therapy against cancer: tumoral suppression induced by glutaminase dysregulation. Spanish Ministry of Science.
- GEM20: Determination of genetic and epigenetic variants associated with response to immune checkpoint inhibitors in cancer. Best Melanoma Research Project.
- COV20-62050: Identification of susceptibility factors for SARS-CoV-2 infection and severity of COVID19 in patients treated with Immune Checkpoint Blockade. Andalusian Government, Science.
- PI-0121-2020: Prospective longitudinal study of genetic, epigenetic and cell populations markers of response to Immune Checkpoint Blockade in melanoma. Andalusian Government, Health.
Guardamagna M, Meyer ML, Berciano-Guerrero MÁ, Mesas-Ruiz A, Cobo-Dols M, Perez-Ruiz E, Cantero Gonzalez A, Lavado-Valenzuela R, Barragán I, Oliver J, Garrido-Aranda A, Alvarez M, Rueda-Dominguez A, Queipo-Ortuño MI, Alba Conejo E, Benitez JC. Oncogene-addicted solid tumors and microbiome-lung cancer as a main character: a narrative review. Transl Lung Cancer Res. 2024 Aug 31;13(8):2050-2066. doi: 10.21037/tlcr-24-216. Epub 2024 Aug 6. PMID: 39263011 Free PMC article. Review.
Zafra J, Onieva JL, Oliver J, Garrido-Barros M, González-Hernández A, Martínez-Gálvez B, Román A, Ordóñez-Marmolejo R, Pérez-Ruiz E, Benítez JC, Mesas A, Vera A, Chicas-Sett R, Rueda-Domínguez A, Barragán I. Novel Blood Biomarkers for Response Prediction and Monitoring of Stereotactic Ablative Radiotherapy and Immunotherapy in Metastatic Oligoprogressive Lung Cancer. Int J Mol Sci. 2024 Apr 20;25(8):4533. doi: 10.3390/ijms25084533. PMID: 38674117 Free PMC article.
Omotesho QA, Escamilla A, Pérez-Ruiz E, Frecha CA, Rueda-Domínguez A, Barragán I. Epigenetic targets to enhance antitumor immune response through the induction of tertiary lymphoid structures. Front Immunol. 2024 Jan 25;15:1348156. doi: 10.3389/fimmu.2024.1348156. eCollection 2024. PMID: 38333212 Free PMC article. Review.
Oliver J, Onieva JL, Garrido-Barros M, Cobo-Dols M, Martínez-Gálvez B, García-Pelícano AI, Dubbelman J, Benítez JC, Martín JZ, Cantero A, Perez-Ruiz E, Rueda-Dominguez A, Barragan I. Fluorometric Quantification of Total Cell-Free DNA as a Prognostic Biomarker in Non-Small-Cell Lung Cancer Patients Treated with Immune Checkpoint Blockade. Cancers 2023, 15, 3357. https:// doi.org/10.3390/cancers15133357
Chaves P, Garrido M, Oliver J, Pérez-Ruiz E, Barragan I, Rueda-Domínguez A. Preclinical models in head and neck squamous cell carcinoma. Br J Cancer. 2023 May;128(10):1819-1827. doi: 10.1038/s41416-023-02186-1. Epub 2023 Feb 10. Review. PubMed PMID: 36765175; PubMed Central PMCID: PMC10147614.
Chicas-Sett R, Zafra J, Rodriguez-Abreu D, Castilla-Martinez J, Benitez G, Salas B, Hernandez S, Lloret M, Onieva JL, Barragan I, Lara PC. Combination of SABR With Anti-PD-1 in Oligoprogressive Non-Small Cell Lung Cancer and Melanoma: Results of a Prospective Multicenter Observational Study. Int J Radiat Oncol Biol Phys. 2022 Nov 15;114(4):655-665. doi: 10.1016/j.ijrobp.2022.05.013. Epub 2022 May 18. PubMed PMID: 35595158.
García Aranda M, López-Rodríguez I, García-Gutiérrez S, Padilla-Ruiz M, de Luque V, Hortas ML, Diaz T, Álvarez M, Barragan-Mallofret I, Redondo M; Research Network on Health Services in Chronic Diseases (Red de Investigación en Servicios Sanitarios y Enfermedades Crónicas, REDISSEC). Laboratory protocol for the digital multiplexed gene expression analysis of nasopharyngeal swab samples using the NanoString nCounter system. F1000Res. 2022 Feb 2;11:133. doi: 10.12688/f1000research.103533.2. eCollection 2022.PMID: 36329793 Free PMC article.
Guardamagna M, Berciano-Guerrero MA, Villaescusa-González B, Perez-Ruiz E, Oliver J, Lavado-Valenzuela R, Rueda-Dominguez A, Barragán I, Queipo-Ortuño MI. Gut Microbiota and Therapy in Metastatic Melanoma: Focus on MAPK Pathway Inhibition. Int J Mol Sci. 2022 Oct 9;23(19). doi: 10.3390/ijms231911990. Review. PubMed PMID: 36233289; PubMed Central PMCID: PMC9569448.
Oliver J, Onieva JL, Garrido-Barros M, Berciano-Guerrero MÁ, Sánchez-Muñoz A, José Lozano M, Farngren A, Álvarez M, Martínez-Gálvez B, Pérez-Ruiz E, Alba E, Cobo M, Rueda-Domínguez A, Barragán I. Association of Circular RNA and Long Non-Coding RNA Dysregulation with the Clinical Response to Immune Checkpoint Blockade in Cutaneous Metastatic Melanoma. Biomedicines. 2022 Sep 27;10(10). doi: 10.3390/biomedicines10102419. PubMed PMID: 36289681; PubMed Central PMCID: PMC9599084.
Berciano-Guerrero MA, Guardamagna M, Perez-Ruiz E, Jurado JM, Barragán I, Rueda-Dominguez A. Treatment of Metastatic Melanoma at First Diagnosis: Review of the Literature. Life (Basel). 2022 Aug 24;12(9). doi: 10.3390/life12091302. Review. PubMed PMID: 36143339; PubMed Central PMCID: PMC9505710.
Onieva JL, Xiao Q, Berciano-Guerrero MÁ, Laborda-Illanes A, de Andrea C, Chaves P, Piñeiro P, Garrido-Aranda A, Gallego E, Sojo B, Gálvez L, Chica-Parrado R, Prieto D, Pérez-Ruiz E, Farngren A, Lozano MJ, Álvarez M, Jiménez P, Sánchez-Muñoz A, Oliver J, Cobo M, Alba E, Barragán I. High IGKC-Expressing Intratumoral Plasma Cells Predict Response to Immune Checkpoint Blockade. Int J Mol Sci. 2022 Aug 15;23(16). doi: 10.3390/ijms23169124. PubMed PMID: 36012390; PubMed Central PMCID: PMC9408876.
Oliver J, Garcia-Aranda M, Chaves P, Alba E, Cobo-Dols M, Onieva JL, Barragan I. Emerging non-invasive methylation biomarkers of cancer prognosis and drug response prediction. Semin Cancer Biol. 2022 Aug;83:584-595. doi: 10.1016/j.semcancer.2021.03.012. Epub 2021 Mar 20. Review. PubMed PMID: 33757849.
Søndergaard JN, Sommerauer C, Atanasoai I, Hinte LC, Geng K, Guiducci G, Bräutigam L, Aouadi M, Stojic L, Barragan I, Kutter C. CCT3-LINC00326 axis regulates hepatocarcinogenic lipid metabolism. Gut. 2022 Jan 12;. doi: 10.1136/gutjnl-2021-325109. [Epub ahead of print] PubMed PMID: 35022268; PubMed Central PMCID: PMC9484377.
Pérez-Ruiz E, Gutiérrez V, Muñoz M, Oliver J, Sánchez M, Gálvez-Carvajal L, Rueda-Domínguez A, Barragán I. Liquid Biopsy as a Tool for the Characterisation and Early Detection of the Field Cancerization Effect in Patients with Oral Cavity Carcinoma. Biomedicines. 2021 Oct 15;9(10). doi: 10.3390/biomedicines9101478. Review. PubMed PMID: 34680596; PubMed Central PMCID: PMC8533108.
Barragan I. Epigenetics modulates the complexity of the response to Immune Checkpoint Blockade. EBioMedicine. 2020 Oct;60:103005. doi: 10.1016/j.ebiom.2020.103005. Epub 2020 Sep 26. PubMed PMID: 32987318; PubMed Central PMCID: PMC7522086.
Gasca J, Flores ML, Jiménez-Guerrero R, Sáez ME, Barragán I, Ruíz-Borrego M, Tortolero M, Romero F, Sáez C, Japón MA. EDIL3 promotes epithelial-mesenchymal transition and paclitaxel resistance through its interaction with integrin αVβ3 in cancer cells. Cell Death Discov. 2020;6:86. doi: 10.1038/s41420-020-00322-x. eCollection 2020. PubMed PMID: 33014430; PubMed Central PMCID: PMC7494865.
Chica-Parrado MR, Godoy-Ortiz A, Jiménez B, Ribelles N, Barragan I, Alba E. Resistance to Neoadjuvant Treatment in Breast Cancer: Clinicopathological and Molecular Predictors. Cancers (Basel). 2020 Jul 22;12(8). doi: 10.3390/cancers12082012. Review. PubMed PMID: 32708049; PubMed Central PMCID: PMC7463925.
Xiao Q, Nobre A, Piñeiro P, Berciano-Guerrero MÁ, Alba E, Cobo M, Lauschke VM, Barragán I. Genetic and Epigenetic Biomarkers of Immune Checkpoint Blockade Response. J Clin Med. 2020 Jan 20;9(1). doi: 10.3390/jcm9010286. Review. PubMed PMID: 31968651; PubMed Central PMCID: PMC7019273.
Lauschke VM, Nordling Å, Zhou Y, Fontalva S, Barragan I, Ingelman-Sundberg M. CYP3A5 is unlikely to mediate anticancer drug resistance in hepatocellular carcinoma. Pharmacogenomics. 2019 Oct;20(15):1085-1092. doi: 10.2217/pgs-2019-0094. PubMed PMID: 31588878.