Our research
Open Epigenetic origins and mechanisms in neuroinflammation configuration options
Our vision is to explain how epigenome integrates instructions from genetic and lifestyle factors rendering immune cells aggressive and brain cells vulnerable in persons affected by Multiple Sclerosis.
Our goal is to better understand disease etiology and to improve disease management through novel personalized treatments and more specific and sensitive biomarkers.
We are a highly cooperative international group committed to spearhead translational epigenetics research. We are committed to answering pertinent clinical questions by combining deep molecular profiling in unique clinical materials using cutting-edge tools with advanced functional experimental models.

Epigenetic origins and mechanisms in neuroinflammation
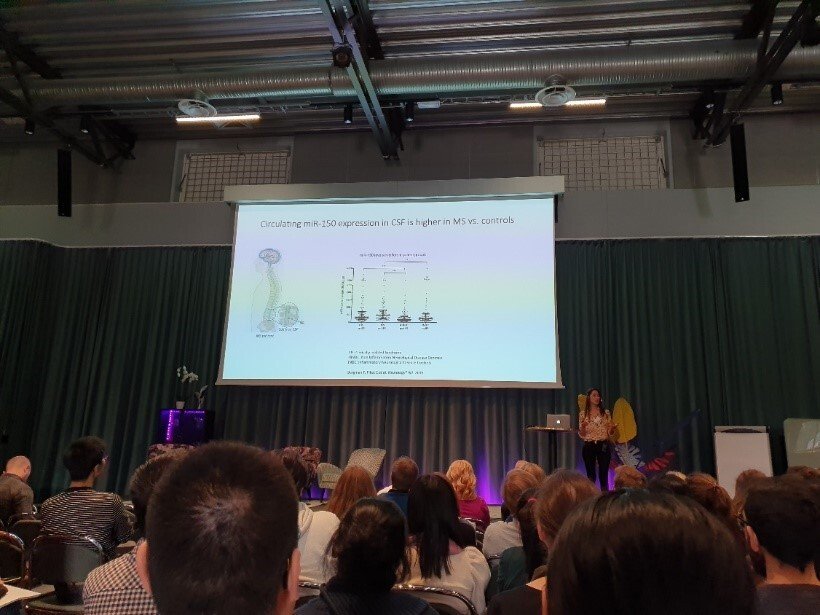
Multiple Sclerosis (MS) is a chronic inflammatory disease characterized by autoimmune destruction of myelin and neurons in the central nervous system. Today, MS is the most common cause of non-traumatic neurological disability among young adults. Predisposition to MS, similar to other common diseases, irrefutably depends on the complex interplay between genetic and environmental factors. Nevertheless, the epigenetic mechanisms that provide a molecular link between the genome and ‘environmental’ signals and control activity of the genome are still virtually unexplored.
Epigenetic changes are heritable through cell division, controlling gene expression without altering DNA sequence (the genetic code). They provide additional and more flexible level of regulation on the top of the genetic code that can also be modulated by environment. We focus on the role of DNA methylation and non-coding RNAs (ncRNAs), especially microRNAs (miRNA).
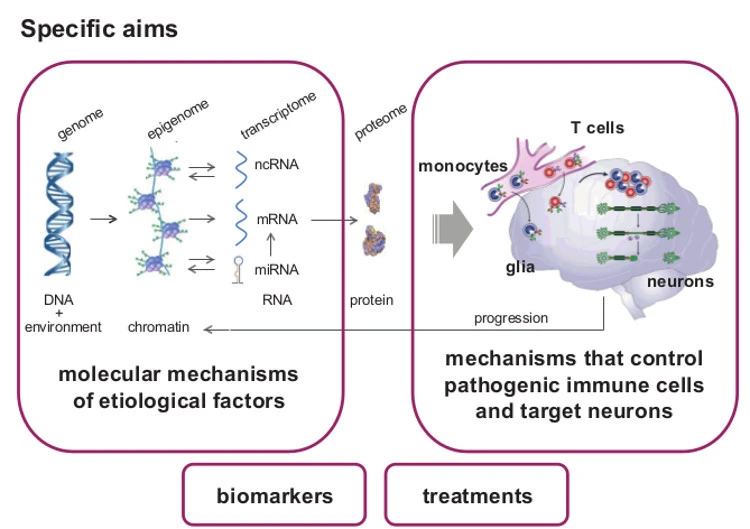
Due to their stability, epigenetic changes may provide better etiologic clues and biomarkers. Due to their reversibility, it will become possible to alter unfavorable epigenetic states towards recovery. Therefore, characterizing epigenetic mechanisms gives tremendous opportunities and may open promising insights into pathogenesis of MS, facilitate diagnosis and improve drug development and the treatment of MS patients.