Our research
The molecular mechanisms that control chromosome dynamics and maintain genome stability are essential for life and prevent accumulation of disease-promoting chromosomal aberrations. With the aim to decipher these mechanisms, our projects focus on the evolutionary conserved family of SMC protein complexes (SMC: Structural Maintenance of Chromosomes) which we investigate using the budding yeast Saccharomyces cerevisiae model organism, and biochemical and structural in vitro analysis.
Considered as an entity, the eukaryotic complexes cohesin, condensin and the Smc5/6 complex control most chromosome-based processes, including replication, segregation, repair and transcription. While it has become increasingly clear that SMC complexes act by structurally organizing chromosomes, their exact modes of action remain unclear. This is especially true for the Smc5/6 complex, which is the main object of investigation in our team.
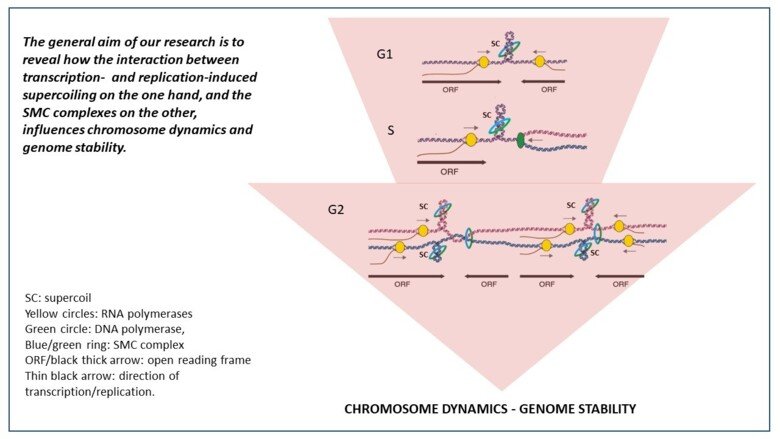
The Smc5/6 complex has mainly been functionally connected with DNA repair and recombination, and we have analyzed this function in both mitotic and meiotic cells. We have also shown that Smc5/6 has a non-repair function, and accumulates on replicated chromosomes in unchallenged cells. Intriguingly, this enrichment increases in linear correlation with the length of the chromosomes, and our investigations show that this is due to a functional connection between the Smc5/6 complex and DNA supercoiling. Supercoiling is the under- or over-twisting of the DNA double helix, and arises when the replication or transcription machineries pry the helix apart. If enzymes called topoisomerases do not remove the supercoils, they inhibit replication and transcription, and increase the risk of genomic instability. Our ongoing investigations aim to understand how transcription- and replication-induced supercoiling influences the function of SMC complexes, chromosome dynamics and genome stability. We also investigate the role of Smc5/6 and supercoiling in the control of Hepatitis B virus and that of DNA supercoiling and topoisomerases in segregation of mitochondrial DNA in a collaborative project headed by Dr. Maria Falkenberg at Gothenburg University.
Perform your master thesis project with us
Please send an email with your CV and motivation letter to camilla.bjorkegren@ki.se if you are interested to perform your master thesis project in our group.

