Tips for New and Experienced Microscope Users
Boost your skills and knowledge in microscopy to get most of your samples and avoid annoying mistakes.
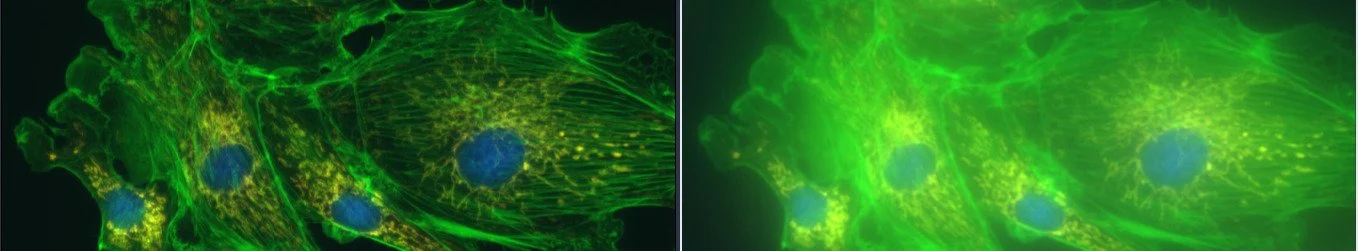
Correct vs incorrect set-up for image acquisition. Images taken by Göran Månsson.
Some tips to get best imaging results
Sample preparation
- Use correct coverslips: For the best imaging results use #1.5 coverslips (0.17mm thickness) as most microscope objectives are optimized for these.
- Coverslip coverage (concerns only inverted microscopes): Avoid placing the coverslip all the way to the end of the slide. This can cause the slide to tilt in the holder because one end will have the combined thickness of the slide and coverslip, while the other end will only have the slide's thickness.
- Multiwell plate imaging: Avoid seeding cells in the edge wells of 96-well and 384-well plates. In most our systems, the stage's travel range is limited to prevent collisions with the objectives.
- Choose right fluorophores: Match your fluorophores to the available laser lines and excitation-emission filters; check the cross-talk between fluorophores by, for example, Fluorescence SpectraViewer.
- Reduce background signal: Wash well your samples, avoid phenol red in the media. For thick, fixed specimens, use clearing methods to reduce light scattering and improve transparency. Quench autofluorescence by exposing to the bright light before staining.
Imaging
- Sample temperature: Let fixed samples to reach room temperature before imaging. Cold samples warm up on the microscope, causing focus drift. Warm up the microscope imaging chamber well before imaging live samples.
- Match the refractive indices of the immersion and embedding media. For example, if you use glycerol-based mounting media like ProLong® (n≈1.47), oil (n ≈ 1.52) or glycerol (n ≈1.47) objectives would be good. For live cells, you will be imaging in cell media (n≈1.33) and therefore should work with water objectives.
- Objective correction collar: Some objectives have a correction collar that can be adjusted to match the sample's bottom thickness. In our systems, the default setting is 0.17mm. Users with samples of different bottom thicknesses must adjust the collar for optimal imaging results. If unsure, ask for help and reset to 0.17mm after use.
- Bit Depth Selection: Choose 8-bit settings for routine imaging and 16-bit for high-dynamic-range imaging to capture more details or weak images.
- Balance between light intensity and signal amplification: High light intensity can bleach your image and harm live cells. Start by amplifying your signal with detector gain or camera exposure. If these settings get too high (causing increased noise or long acquisition times), then increase the light intensity. If possible, adjust imaging settings on non-important areas.
- Speed up acquisition: Crop the scanning area to focus only on the region of interest. This reduces the number of pixels and speeds up acquisition. Do not exceed the optimal X/Y and Z-stack settings suggested by the software to avoid collecting unnecessary data without improving image quality. For fixed samples with multiple channels, scan the entire 3D stack with one channel before switching to the next to speed up image collection.
Post-processing
- Enhance Intensity: Note that sometimes images, especially 16-bit ones, may appear weak in analysis software. Adjust brightness/contrast to improve the visibility. For consistency, adjust one image and apply the same values to the rest of your experimental images.
- Enhance Resolution: Apply deblurring techniques and deconvolution algorithms to enhance resolution and reduce background.
- Colours on your images: Avoid using blue on a black background – our eye struggles to differentiate this combination. Opt for grayscale to improve the visibility of weak signals. When presenting multicolour images, it's considerate to avoid using green and red together. Instead, use green and magenta, or consider cyan, yellow, and magenta color scheme.
How to clean the microscope objectives
BIC_How to clean microscope objectives (PDF, 659.99 KB)
Resources for image collection and analysis
We collected here some useful software with tutorials and examples that can be helpful for your image analysis.
- Fiji/ImageJ is an open-source software for microscope image analysis. With Fiji, you can visualize your results, utilize custom plugins, and create your own analysis macros for automated image processing and quantification. Explore the ImageJ tutorials to get started with image analysis and enhance your skills.
- CellProfiler is a free software dedicated to cell image analysis. It allows you to design pipelines for automatic processing and measure the phenotypes of interest in your images. Follow online CellProfiler tutorials and look at CellProfiler examples to learn how to build your own analysis pipelines. BIC users can also request access to materials from the CellProfiler workshop held by BIC.
- Imaris, a powerful 3D and 4D image analysis software, enables scientists to explore complex biological data, visualize cellular structures, track dynamic processes, and quantify features within multidimensional microscopy images. Check out the collections of tutorial videos (1, 2, 3) to get the most of Imaris’ capabilities.
- The Microscopy Series on iBiology is a collection of free online tutorials that cover the basics of optics, transmitted light microscopy, various methods of imaging fluorescent samples, camera operation, image processing, and the latest advances in light microscopy.
- MicroscopyU, created by Nikon, is an educational resource that offers comprehensive information, interactive tutorials, and timely updates on all aspects of optical microscopy, photomicrography, and digital imaging.
- The Fluorescence SpectraViewer by Thermo Fisher Scientific is a tool that allows users to compare the excitation and emission parameters for many fluorophores, helping them find compatible reagents and optimal filters for their experiments.
- Euro-BioImaging is a European research infrastructure that provides open access to state-of-the-art imaging technologies, training, and data services in biological and biomedical imaging for life science researchers.
