Our research
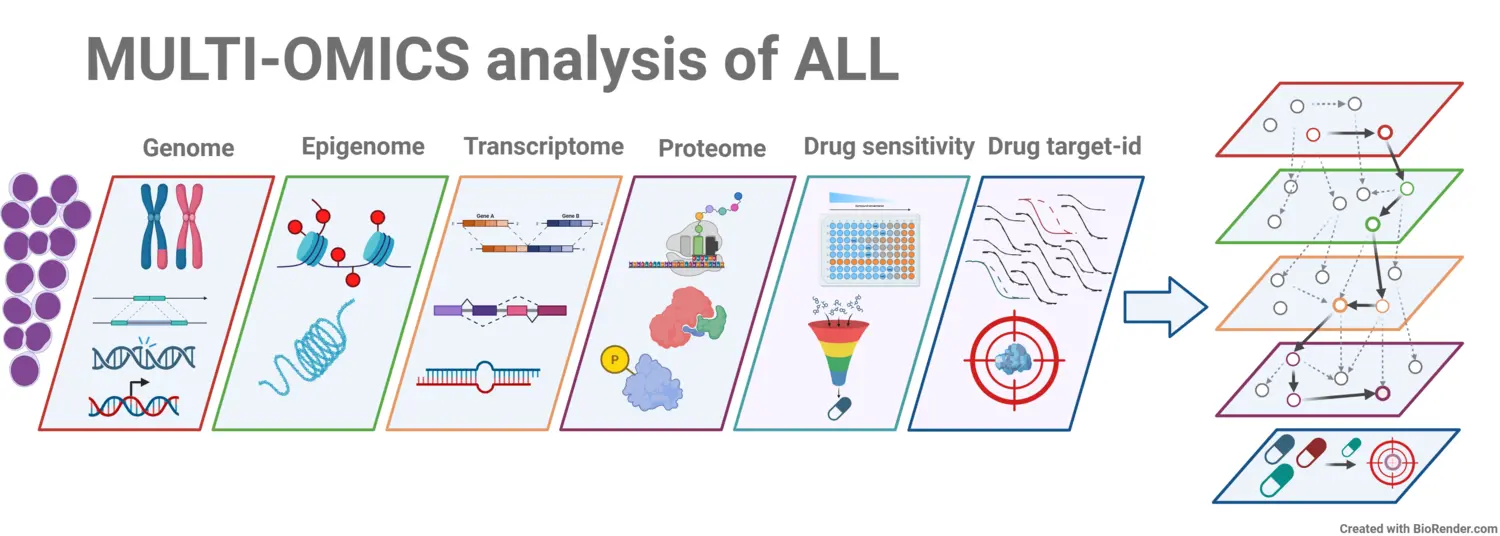
We are utilizing and integrating state-of-the-art omics methods to obtain detailed molecular phenotypes of childhood and adult acute lymphoblastic leukemia (ALL), aiming to identify precision medicine candidates for improved therapy. Building on our recent studies (Leo, I.R. et al., Nat Commun 13, 1691 (2022), PMID:35354797, Kurzawa, N. et al., Nat Chem Biol 19, 962–971 (2023), PMID:36941476), we are advancing the study and validation of precision medicine candidates, with a strong focus on their translational implementation.
A fundamental question we are currently addressing is how cancer genome aberrations influence the functional proteome by combining genomics with proteomics data, a field known as cancer proteogenomics. In addition to quantitative proteomics, we are enhancing our analysis by capturing the biophysical state of the proteome, regardless of its abundance, using thermal proteomics. This approach allows us to identify functional proteoforms as well as their association with drug sensitivity.
Furthermore, we are investigating the target landscape of current and emerging targeted therapeutics using orthogonal chemical proteomics approaches to understand their mechanisms of action and potential toxicities. Our research also focuses on molecular and cellular biology, aiming to translate our findings into clinical applications that can significantly improve patient outcomes.