
Personalized Medicine and Drug Development – Lauschke-lab
We integrate 3D cell culture systems of primary human cells, microfluidics and comprehensive molecular profiling technologies to discover novel therapeutic strategies for inflammatory conditions (NASH), infectious diseases (COVID-19 and hemorrhagic fevers) and complex metabolic diseases (type 2 diabetes).
Our Research
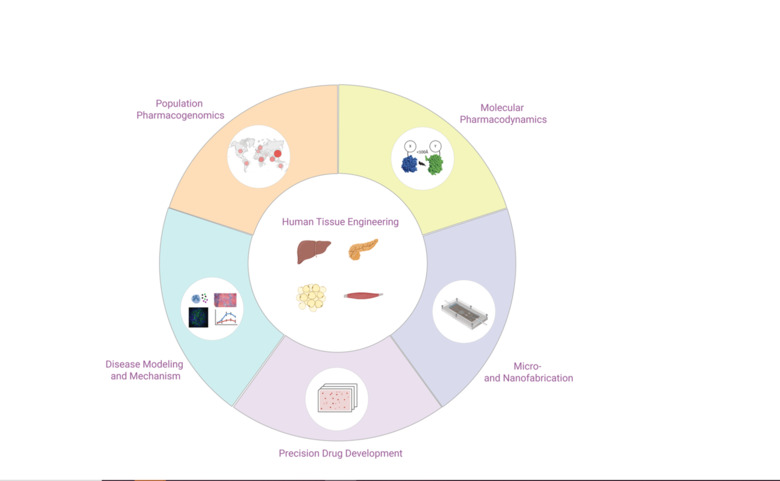
We integrate 3D cell culture systems of primary human cells, microfluidics and comprehensive molecular profiling technologies to discover novel therapeutic strategies for inflammatory conditions (NASH), infectious diseases (COVID-19 and hemorrhagic fevers) and complex metabolic diseases (type 2 diabetes).
In addition, we use population-scale genetics and machine learning tools to map the ethnogeographic variability in genes involved in drug pharmacokinetics and pharmacodynamics with the goal to improve personalized medicine and precision public health.
Background
The number of successful drug development projects has stagnated for decades, despite major breakthroughs in chemistry, molecular biology and genetics. Unreliable target identification and poor translatability of preclinical results have been identified as major causes of failure. To improve predictions of clinical efficacy and safety, interest has shifted from conventional 2D cultures to organotypic and microphysiological culture methods in which human cells can retain physiologically and functionally relevant phenotypes for extended periods of time. The Lauschke lab develops 3D cell culture models of primary human tissues and microfluidic devices for the discovery and development. By interfacing these systems with high-throughput screening, we aim to develop novel drug candidates for a variety of indications, including NASH, type 2 diabetes, COVID-19 and hemorrhagic fevers.
In addition, we integrate population-scale genomics with machine learning to optimize pharmacogenomic drug response predictions and facilitate the implementation of NGS in personalized medicine and precision public health.
Projects
Bioengineering of 3D primary human tissue models
We develop and characterize long-term stable 3D human tissue models of liver, pancreas, adipose tissue and skeletal muscle. Importantly, we exclusively use patient-derived cells, i.e. no cell lines or stem cells, and integration of histological, transcriptomic, proteomic and metabolomic signatures demonstrates that the tissue models feature mature phenotypes that facilitate improved result translation. A main area of our interests is the integration of different cell types (endothelial cells, immune cells, etc.) and the functional characterization of cell-cell interactions in both health and disease.
Micro and Nanofabrication for biomedical applications
Using state-of-the-art cleanroom tools, we leverage nanofabrication technologies for their implementation in biomedicine. Uniquely, we have established a novel nanofabrication platform, termed Nano Reaction Injection Molding (NanoRIM) for the rapid, versatile and cost-effective fabrication of polymer micro- and nanodevices. We further utilize micro- and nanopatterned surfaces to study the interaction of human cells with surface topographies.
We employ diverse materials, including polymers and inorganics, such as silicon and glass, with appropriate surface treatment for different biomedical applications. Specifically, we develop platforms for pharmacological and toxicological applications using materials with minimal drug absorption.
Development of microfluidic multi-organ co-culture chips
In the field of drug development and discovery, the poor predictive accuracy of preclinical models due to pronounced species differences has led to a high rate of project attrition. The co-culturing of human cells from different tissues and the subsequent tissue-tissue interaction in perfused microfluidic devices have shown great potential to tackle this problem in ex vivo.
By integrating our custom-designed microfluidic systems with 3D human tissue models, we have developed microphysiological organ-on-a-chip platforms with superior function that allow to mimic tissue-tissue interactions. Specifically, we integrate human 3D tissue models of liver, pancreas, adipose tissue and skeletal muscle, and utilize this platform as a model of integrative glycemic control.
Monitoring of intracellular signal transduction in primary human tissues
We monitor signaling of G protein-coupled receptors (GPCRs) and other signal transduction events in primary human tissue models using highly sensitive and selective bioluminescence resonance energy transfer (BRET)-based biosensors. The obtained information sheds new light on the functional selectivity and subcellular signaling of important signaling cascades without the need to overexpress the receptors. Employing such biosensor-based approaches to map signaling events in fine detail holds great promise for our understanding of drug efficacy and side effects. Combining these tools with patient-derived material will provide much needed insight into the intra- and inter-patient variability of drug action and offers the potential to develop better and more targeted therapies.
Hepatic spheroids as model system for liver regeneration
Strikingly, during spheroid aggregation stages, hepatocytes first dedifferentiate, followed by rapid redifferentiation, providing an ideal ex vivo experimental paradigm to study the full spectrum of differentiation state changes that occur in vivo during liver regeneration. Besides extending our mechanistic understanding, this finding opened possibilities for the development of therapeutic approaches as a substitute for orthotopic liver transplantations. We work on the establishment of protocols in which hepatocytes isolated from patients proliferate and, after cells sufficiently multiplied, are induced to redifferentiate into functional hepatocytes using our 3D spheroid culture system. Furthermore, we investigate the molecular mechanisms underlying liver hypertrophy and develop drug candidates that improve hepatocyte proliferation.
Evaluation of the importance of rare genetic variants on hepatic metabolism and drug response
Genetic variants primarily in drug and metabolite transporters, phase I and phase II drug metabolizing enzymes and nuclear receptors can influence drug response by modulating drug absorption, distribution, metabolism and excretion (ADME). Importantly, while in the past decades an ever-growing arsenal of genetic variants with demonstrated impacts on human drug response has been identified in these pharmacogenes, a substantial fraction of the heritable variability in drug response remains unexplained. Rare genetic variants that only occur in very few individuals and are hence missed in genome-wide association studies have been proposed to contribute to this missing heritability.
We integrate data from recent population-wide Next-Generation Sequencing (NGS) projects to quantify the extent of genetic variability in pharmacogenes on a population level and, using an arsenal of in silico techniques, quantify the impact on hepatic metabolism and pharmacokinetics and -dynamics.
Publications
Selected publications
- Article: BJA OPEN. 2023;7:100218Widaeus M; Hertzberg D; Hallqvist L; Bell M
- Article: BMJ OPEN. 2023;13(4):e071135Lindeberg FCB; Bell M; Larsson E; Hallqvist L
- Article: EUROPEAN JOURNAL OF EPIDEMIOLOGY (EJE). 2023;38(3):301-311Bell M; Ekbom A; Linder M
- Article: BMC MEDICINE. 2023;21(1):1Bell M; Hergens M-P; Fors S; Tynelius P; de Leon AP; Lager A
- Article: ACTA ANAESTHESIOLOGICA SCANDINAVICA. 2022;66(10):1185-1192Valadkhani A; Henningsson R; Nordstrom JL; Granstrom A; Hallqvist L; Wahlgren CM; Peterzen B; Eriksson J; Bell M; Gupta A
- Article: MEDICAL JOURNAL OF AUSTRALIA. 2022;217(6):311-317Reilly JR; Myles PS; Wong D; Heritier SR; Brown WA; Richards T; Bell M
- Article: EUROPEAN JOURNAL OF EPIDEMIOLOGY (EJE). 2022;37(2):157-165Hergens M-P; Bell M; Haglund P; Sundstrom J; Lampa E; Nederby-Ohd J; Ostlund MR; Cars T
- Article: CLINICAL MEDICINE INSIGHTS: CARDIOLOGY. 2022;16:11795468221133611Moore A; Bell M
- Article: BLOOD PURIFICATION. 2022;51(7):584-589Hertzberg D; Renberg M; Nyman J; Bell M; Stigare CR
- Article: GASTROENTEROLOGY. 2021;161(6):1982-1997.e11Azzimato V; Chen P; Barreby E; Morgantini C; Levi L; Vankova A; Jager J; Sulen A; Diotallevi M; Shen JX; Miller A; Ellis E; Ryden M; Naslund E; Thorell A; Lauschke VM; Channon KM; Crabtree MJ; Haschemi A; Craige SM; Mori M; Spallotta F; Aouadi M
- Article: PLOS MEDICINE. 2021;18(10):e1003820Bergqvist R; Ahlqvist VH; Lundberg M; Hergens M-P; Sundstrom J; Bell M; Magnusson C
- Article: SCIENCE ADVANCES. 2021;7(36):eabi6856Zhou Y; Arribas GH; Turku A; Juergenson T; Mkrtchian S; Krebs K; Wang Y; Svobodova B; Milani L; Schulte G; Korabecny J; Gastaldello S; Lauschke VM
- Article: JOURNAL OF ELECTROCARDIOLOGY. 2021;68:1-5Sapra R; Hallqvist L; Schlegel TT; Ugander M; Bell M; Maanja M
- Article: ADVANCED SCIENCE. 2021;8(16):e2100106Shen JX; Couchet M; Dufau J; de Castro Barbosa T; Ulbrich MH; Helmstadter M; Kemas AM; Zandi Shafagh R; Marques M-A; Hansen JB; Mejhert N; Langin D; Ryden M; Lauschke VM
- Editorial comment: NEW ENGLAND JOURNAL OF MEDICINE. 2021;385(5):463-465Stebbing J; Lauschke VM
- Article: EMBO MOLECULAR MEDICINE. 2020;12(8):e12697Stebbing J; Krishnan V; de Bono S; Ottaviani S; Casalini G; Richardson PJ; Monteil V; Lauschke VM; Mirazimi A; Youhanna S; Tan Y-J; Baldanti F; Sarasini A; Terres JAR; Nickoloff BJ; Higgs RE; Rocha G; Byers NL; Schlichting DE; Nirula A; Cardoso A; Corbellino M
- Article: ADVANCED SCIENCE. 2020;7(15):2000248Oliva-Vilarnau N; Vorrink SU; Ingelman-Sundberg M; Lauschke VM
- Article: SCIENCE TRANSLATIONAL MEDICINE. 2020;12(532):eaaw9709Azzimato V; Jager J; Chen P; Morgantini C; Levi L; Barreby E; Sulen A; Oses C; Willerbrords J; Xu C; Li X; Shen JX; Akbar N; Haag L; Ellis E; Walhen K; Naslund E; Thorell A; Choudhury RP; Lauschke VM; Ryden M; Craige SM; Aouadi M
- Article: CELL. 2018;172(5):1079-1090.e12Sonnen KF; Lauschke VM; Uraji J; Falk HJ; Petersen Y; Funk MC; Beaupeux M; Francois P; Merten CA; Aulehla A
- Article: NATURE. 2013;493(7430):101-105Lauschke VM; Tsiairis CD; Francois P; Aulehla A
Funding
- Swedish Research Council
- Innovative Medicines Initiative-2 (IMI2) Project EUbOPEN
- Knut and Alice Wallenberg Foundation
- Organ-On-A-Chip Technologies Network Grant
- Horizon 2020 program of the European Union (U-PGx)
In addition, the group acknowledges generous support from the Robert Bosch Foundation for Medical Research (RBMF), Karolinska Institutet, Eli Lilly and Company and Merck KGaA.
Staff and contact
Group leader
- Volker LauschkeProfessor | Docent
All members of the group
- Isabel BarraganAffiliated to Research
- Mahamadou CamaraPhd Student
- Aurino KemasPhd Student
- Jibbe KeulenAffiliated to Research
- Stefania KoutsilieriPhd Student
- Volker LauschkeProfessor | Docent
- Xuexin LiAffiliated to Research
- Evgeniya MickolsAffiliated to Research
- Yoomi ParkAffiliated to Research
- Yvonne PlattnerAffiliated to Research
- Nayere TaebniaPostdoctoral Researcher
- Sabine WillemsPostdoctoral Studies
- Shane WrightPostdoctoral Researcher | Assistant Professor
- Chen XingPhd Student
- Sonia YouhannaAffiliated to Research
- Reza Zandi ShafaghResearch Specialist
- Yitian ZhouPostdoctoral Researcher
Pharmacoepigenetics

Our ultimate goal is to understand the pharmacological response and identify new biomarkers that are amenable for targeted manipulation. Our strategy is to combine genetic and epigenetic state-of-the-art analyses of clinical samples and functional studies in advanced three-dimensional cell cultures.
We developed a method that distinguishes DNA methylation from hydroxymethylation, and identified variants specific to each of these cytosine modifications important for drug metabolism (Nucleic Acids Research, 2016).
We have recently applied this principle to the identification of the first epigenetic marker of response to cancer immunotherapy (Lancer Resp. Med., 2018, Best Practice, 2019). In addition, we have identified a novel transcriptomic signature of B cell functionality associated with survival in melanoma patients treated with immunotherapy (IJMS 2023).
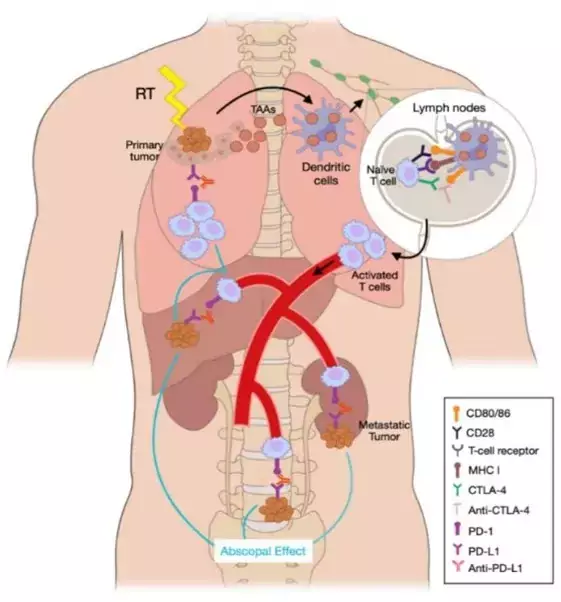
Another interesting aspect of our research is Abscopal Response, a distant response in a non-irradiated tumor after radiotherapy in other lesions. We have reported a 65% Abscopal Response rate that was associated with an improved Progression-Free Survival (21.2 months vs 3 months, p < 0.0001) in patients that were treated with the combination of Radiotherapy and Immunotherapy (International Journal of Radiation Oncology, 2022).
Our recent projects incorporate this combination therapy with the aim of characterizing the immunogenic cell death that Immune Checkpoint Blockade triggers in those patients developing Abscopal Response. In a preliminary cohort, we have correlated a high percentage of CD4+ and CD8+PDL1 and a low percentage of CD8+PD1+ with a good response. Longitudinally, we also found that an increase in circulating free DNA (cfDNA) concentration during SABR related to a poor response, whereas an increase in CD20+ cells and a decrease in CD19+ cells after the first SABR fraction were associated with a favorable response. Furthermore, we have identified a set of microRNAs (miRNAs) in patients with non-small cell lung cancer that are predictive of response (Zafra J et al., 2024).
Specialised methods
Chicas-sett R et al. Cancers (Basel). 2020;12(8):2178–98.
- Three-dimensional cultures (and treatment/phenotyping) of primary human liver cells and cancer cell lines.
- High-throughput 3D cultures in 96-well plates using hanging drop technology (© InSphero) or Ultra Low Attachment surfaces.
- TET-assisted bisulfite conversion (TAB)-based determination of DNA methylation and hydroxymethylation:
- TAB-EPIC Illumina DNA methylation arrays for genome-wide analysis of 850K CpG sites
- TAB-Methyl-Seq Agilent targeted libraries for Next Generation Sequencing
- TAB-Sanger sequencing
- Adaptations of the protocol for FFPE and cell-free DNA
- TET-independent restriction-based determination of DNA methylation and hydroxymethylation
- Quantification of global DNA methylation and hydroxymethylation by mass spectrometry/liquid chromatography.
- Smart-seq library preparation for single cell RNA-sequencing.
- All RNA-sequencing by ribosomal depletion
- Exome sequencing with DNA from FFPE tumors and paired PBMCs
- ChIP-qPCR for histone modifications in 3D liver cancer cell lines and primary hepatocyte microtissues.
- Bioinformatics for genome-wide and customised transcriptomic/epigenomic analyses. OMICS integration. AI-based prediction models.
- Deciphering the EPIgenetic Signatures of the ABscopal Response towards precision immunotherapy (EPI-SABR, PRYES247250BARR). PI: M Isabel Barragan Mallofret. (IBIMA-Plataforma BIONAND). Spanish Association Against Cancer (AECC). 01/12/2024-30/11/2027. 150.000 €.
- EPIPREDICT: Development of a panel of epigenetic markers in liquid biopsy for prediction of response and prognosis in the context of novel immunotherapy approaches. (DTS23/00114). Instituto de Salud Carlos III. PI: M Isabel Barragán Mallofret (IBIMA-Plataforma BIONAND). 01/01/2024- 31/12/2026. 238.480 €.
- MICROMICS: Ex vivo characterization of the mechanisms associated with Abscopal Response (GEM 2023). PI: M Isabel Barragán Mallofret (IBIMA-Plataforma BIONAND). Melanoma Spanish Group. 01/01/2024-31/12/2025. 40.000 €.
- Multi-modal and Multi-scale Modeling based on Artificial Intelligence Pathways and Spatial Transcriptomics to reveal Lung Cancer Distant Relapse predictors (MAPS-LUNG) (CNS2023-145629). PI: M Isabel Barragán Mallofret (IBIMA-Plataforma BIONAND). Ministry of Science, Innovation and Universities. 01/01/2024-31/12/2025. 399.605 €.
- STINT China-Sweden: Identification of DNA methylation and hydroxymethylation variation and molecular network in primary central nervous system lymphoma
- PI22/01816: Immunotherapy resistance biomarker: integrating Abscopal Response and epigenetics in a non-invasive precision medicine strategy. Health Institute Carlos III.
- ProyExcel_01002: Precision pharmacogenomics of immunotherapy treatment of lung and head and neck cancer. Andalusian Government, Science.
- PI22/01816: Immunotherapy resistance biomarker: integrating Abscopal Response and epigenetics in a non-invasive precision medicine strategy. Health Institute Carlos III.
- SEOM21: MicroRNA signatures in the early detection of the field cancerization in squamous cell oral cavity carcinoma.
- AECC 2020: Radio-immunotherapy in metastatic cancer.
- P20-01326: ORIGEN: Early detection of liver metastases in breast cancer based on the study of methylation and hydroximetylation patterns in the circulating free DNA in blood. Andalusian Government, Science.
- PI18/01592: Omics integration for precision cancer immunotherapy. Health Institute Carlos III.
- Metabolic therapy against cancer: tumoral suppression induced by glutaminase dysregulation. Spanish Ministry of Science.
- GEM20: Determination of genetic and epigenetic variants associated with response to immune checkpoint inhibitors in cancer. Best Melanoma Research Project.
- COV20-62050: Identification of susceptibility factors for SARS-CoV-2 infection and severity of COVID19 in patients treated with Immune Checkpoint Blockade. Andalusian Government, Science.
- PI-0121-2020: Prospective longitudinal study of genetic, epigenetic and cell populations markers of response to Immune Checkpoint Blockade in melanoma. Andalusian Government, Health.
Guardamagna M, Meyer ML, Berciano-Guerrero MÁ, Mesas-Ruiz A, Cobo-Dols M, Perez-Ruiz E, Cantero Gonzalez A, Lavado-Valenzuela R, Barragán I, Oliver J, Garrido-Aranda A, Alvarez M, Rueda-Dominguez A, Queipo-Ortuño MI, Alba Conejo E, Benitez JC. Oncogene-addicted solid tumors and microbiome-lung cancer as a main character: a narrative review. Transl Lung Cancer Res. 2024 Aug 31;13(8):2050-2066. doi: 10.21037/tlcr-24-216. Epub 2024 Aug 6. PMID: 39263011 Free PMC article. Review.
Zafra J, Onieva JL, Oliver J, Garrido-Barros M, González-Hernández A, Martínez-Gálvez B, Román A, Ordóñez-Marmolejo R, Pérez-Ruiz E, Benítez JC, Mesas A, Vera A, Chicas-Sett R, Rueda-Domínguez A, Barragán I. Novel Blood Biomarkers for Response Prediction and Monitoring of Stereotactic Ablative Radiotherapy and Immunotherapy in Metastatic Oligoprogressive Lung Cancer. Int J Mol Sci. 2024 Apr 20;25(8):4533. doi: 10.3390/ijms25084533. PMID: 38674117 Free PMC article.
Omotesho QA, Escamilla A, Pérez-Ruiz E, Frecha CA, Rueda-Domínguez A, Barragán I. Epigenetic targets to enhance antitumor immune response through the induction of tertiary lymphoid structures. Front Immunol. 2024 Jan 25;15:1348156. doi: 10.3389/fimmu.2024.1348156. eCollection 2024. PMID: 38333212 Free PMC article. Review.
Oliver J, Onieva JL, Garrido-Barros M, Cobo-Dols M, Martínez-Gálvez B, García-Pelícano AI, Dubbelman J, Benítez JC, Martín JZ, Cantero A, Perez-Ruiz E, Rueda-Dominguez A, Barragan I. Fluorometric Quantification of Total Cell-Free DNA as a Prognostic Biomarker in Non-Small-Cell Lung Cancer Patients Treated with Immune Checkpoint Blockade. Cancers 2023, 15, 3357. https:// doi.org/10.3390/cancers15133357
Chaves P, Garrido M, Oliver J, Pérez-Ruiz E, Barragan I, Rueda-Domínguez A. Preclinical models in head and neck squamous cell carcinoma. Br J Cancer. 2023 May;128(10):1819-1827. doi: 10.1038/s41416-023-02186-1. Epub 2023 Feb 10. Review. PubMed PMID: 36765175; PubMed Central PMCID: PMC10147614.
Chicas-Sett R, Zafra J, Rodriguez-Abreu D, Castilla-Martinez J, Benitez G, Salas B, Hernandez S, Lloret M, Onieva JL, Barragan I, Lara PC. Combination of SABR With Anti-PD-1 in Oligoprogressive Non-Small Cell Lung Cancer and Melanoma: Results of a Prospective Multicenter Observational Study. Int J Radiat Oncol Biol Phys. 2022 Nov 15;114(4):655-665. doi: 10.1016/j.ijrobp.2022.05.013. Epub 2022 May 18. PubMed PMID: 35595158.
García Aranda M, López-Rodríguez I, García-Gutiérrez S, Padilla-Ruiz M, de Luque V, Hortas ML, Diaz T, Álvarez M, Barragan-Mallofret I, Redondo M; Research Network on Health Services in Chronic Diseases (Red de Investigación en Servicios Sanitarios y Enfermedades Crónicas, REDISSEC). Laboratory protocol for the digital multiplexed gene expression analysis of nasopharyngeal swab samples using the NanoString nCounter system. F1000Res. 2022 Feb 2;11:133. doi: 10.12688/f1000research.103533.2. eCollection 2022.PMID: 36329793 Free PMC article.
Guardamagna M, Berciano-Guerrero MA, Villaescusa-González B, Perez-Ruiz E, Oliver J, Lavado-Valenzuela R, Rueda-Dominguez A, Barragán I, Queipo-Ortuño MI. Gut Microbiota and Therapy in Metastatic Melanoma: Focus on MAPK Pathway Inhibition. Int J Mol Sci. 2022 Oct 9;23(19). doi: 10.3390/ijms231911990. Review. PubMed PMID: 36233289; PubMed Central PMCID: PMC9569448.
Oliver J, Onieva JL, Garrido-Barros M, Berciano-Guerrero MÁ, Sánchez-Muñoz A, José Lozano M, Farngren A, Álvarez M, Martínez-Gálvez B, Pérez-Ruiz E, Alba E, Cobo M, Rueda-Domínguez A, Barragán I. Association of Circular RNA and Long Non-Coding RNA Dysregulation with the Clinical Response to Immune Checkpoint Blockade in Cutaneous Metastatic Melanoma. Biomedicines. 2022 Sep 27;10(10). doi: 10.3390/biomedicines10102419. PubMed PMID: 36289681; PubMed Central PMCID: PMC9599084.
Berciano-Guerrero MA, Guardamagna M, Perez-Ruiz E, Jurado JM, Barragán I, Rueda-Dominguez A. Treatment of Metastatic Melanoma at First Diagnosis: Review of the Literature. Life (Basel). 2022 Aug 24;12(9). doi: 10.3390/life12091302. Review. PubMed PMID: 36143339; PubMed Central PMCID: PMC9505710.
Onieva JL, Xiao Q, Berciano-Guerrero MÁ, Laborda-Illanes A, de Andrea C, Chaves P, Piñeiro P, Garrido-Aranda A, Gallego E, Sojo B, Gálvez L, Chica-Parrado R, Prieto D, Pérez-Ruiz E, Farngren A, Lozano MJ, Álvarez M, Jiménez P, Sánchez-Muñoz A, Oliver J, Cobo M, Alba E, Barragán I. High IGKC-Expressing Intratumoral Plasma Cells Predict Response to Immune Checkpoint Blockade. Int J Mol Sci. 2022 Aug 15;23(16). doi: 10.3390/ijms23169124. PubMed PMID: 36012390; PubMed Central PMCID: PMC9408876.
Oliver J, Garcia-Aranda M, Chaves P, Alba E, Cobo-Dols M, Onieva JL, Barragan I. Emerging non-invasive methylation biomarkers of cancer prognosis and drug response prediction. Semin Cancer Biol. 2022 Aug;83:584-595. doi: 10.1016/j.semcancer.2021.03.012. Epub 2021 Mar 20. Review. PubMed PMID: 33757849.
Søndergaard JN, Sommerauer C, Atanasoai I, Hinte LC, Geng K, Guiducci G, Bräutigam L, Aouadi M, Stojic L, Barragan I, Kutter C. CCT3-LINC00326 axis regulates hepatocarcinogenic lipid metabolism. Gut. 2022 Jan 12;. doi: 10.1136/gutjnl-2021-325109. [Epub ahead of print] PubMed PMID: 35022268; PubMed Central PMCID: PMC9484377.
Pérez-Ruiz E, Gutiérrez V, Muñoz M, Oliver J, Sánchez M, Gálvez-Carvajal L, Rueda-Domínguez A, Barragán I. Liquid Biopsy as a Tool for the Characterisation and Early Detection of the Field Cancerization Effect in Patients with Oral Cavity Carcinoma. Biomedicines. 2021 Oct 15;9(10). doi: 10.3390/biomedicines9101478. Review. PubMed PMID: 34680596; PubMed Central PMCID: PMC8533108.
Barragan I. Epigenetics modulates the complexity of the response to Immune Checkpoint Blockade. EBioMedicine. 2020 Oct;60:103005. doi: 10.1016/j.ebiom.2020.103005. Epub 2020 Sep 26. PubMed PMID: 32987318; PubMed Central PMCID: PMC7522086.
Gasca J, Flores ML, Jiménez-Guerrero R, Sáez ME, Barragán I, Ruíz-Borrego M, Tortolero M, Romero F, Sáez C, Japón MA. EDIL3 promotes epithelial-mesenchymal transition and paclitaxel resistance through its interaction with integrin αVβ3 in cancer cells. Cell Death Discov. 2020;6:86. doi: 10.1038/s41420-020-00322-x. eCollection 2020. PubMed PMID: 33014430; PubMed Central PMCID: PMC7494865.
Chica-Parrado MR, Godoy-Ortiz A, Jiménez B, Ribelles N, Barragan I, Alba E. Resistance to Neoadjuvant Treatment in Breast Cancer: Clinicopathological and Molecular Predictors. Cancers (Basel). 2020 Jul 22;12(8). doi: 10.3390/cancers12082012. Review. PubMed PMID: 32708049; PubMed Central PMCID: PMC7463925.
Xiao Q, Nobre A, Piñeiro P, Berciano-Guerrero MÁ, Alba E, Cobo M, Lauschke VM, Barragán I. Genetic and Epigenetic Biomarkers of Immune Checkpoint Blockade Response. J Clin Med. 2020 Jan 20;9(1). doi: 10.3390/jcm9010286. Review. PubMed PMID: 31968651; PubMed Central PMCID: PMC7019273.
Lauschke VM, Nordling Å, Zhou Y, Fontalva S, Barragan I, Ingelman-Sundberg M. CYP3A5 is unlikely to mediate anticancer drug resistance in hepatocellular carcinoma. Pharmacogenomics. 2019 Oct;20(15):1085-1092. doi: 10.2217/pgs-2019-0094. PubMed PMID: 31588878.
Group members
Isabel Barragan – Research group leader
Laura Espino – Postdoc
Andre Nobre – Postdoc
Shady Sayed – Master thesis student