Team Innate immune responses during viral infections
We aim to understand common global interferon-stimulated gene (ISG) responses against different viruses can provide a novel avenue for viral antagonism.
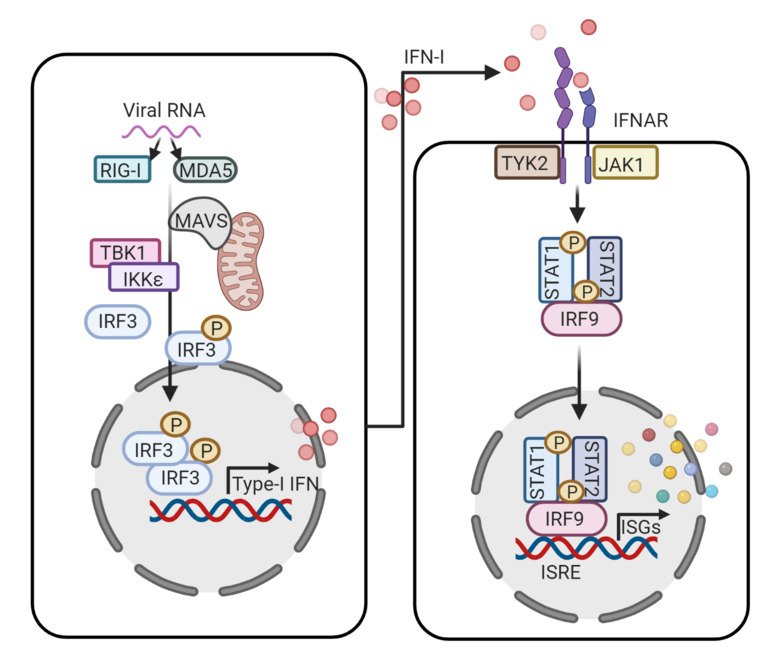
Cell intrinsic and innate immune responses like interferon (IFN) response, autophagy, DNA damage response, etc. play a key role in antiviral defense. Among them, IFN-response serves as the first line of defense. During viral infection or re-activation, the viral nucleic acids and gene products are detected by the cellular pathogen recognition receptors (PRR) like retinoic acid-inducible gene I (RIG-I) and RIG-I like receptors (RLR) like MDA5, lead to activation of a ubiquitin-mediated cascade causing activation of transcription factors IRF3, AP-1, and NF-κB resulting in the production of type-I interferons. The IFN can further activate several ISGs that have anti-viral activities. This is mediated by the binding of IFN-I to its receptors which leads to activation of the JAK-STAT pathway. Humans carry hundreds of ISGs that are induced during viral infection and many of which have direct antiviral activity and can protect against viral infections. Each virus has a unique but partially overlapping antiviral ‘ISG profile’ that can target multiple stages in the virus life cycle. Understanding common global ISG responses against different viruses can provide a novel avenue for viral antagonism.
Interferon-inducible protein ISG15 was one of the first ubiquitin-like modifiers to be identified and is stimulated during infection with several viruses. ISG15 has numerous antiviral functions including inhibition of virus release, ISGylation of both viral and host proteins, and immunomodulatory cytokine-like properties in its unconjugated form. Signaling controlled by ubiquitin or ubiquitin-like proteins such as ISG15 is key regulators of the intrinsic cellular responses. While Ubiquitination is one of the most studied post-translational modifications, not much is known about ISG15 in the context of viral infections. We will identify and characterize antiviral ISGs against different viruses and how the ISG15 landscape is shaped during the virus infection using advanced proteomics technologies and an array of biochemical, siRNA/shRNA knockdown, CRISPR Knockout, and imaging techniques.
Though viruses can elicit a wide array of innate immune responses, most viruses encode proteins that can circumvent host antiviral response by modifying post-translational modifications by ubiquitin or ubiquitin-like proteins such as ISG15, which often alters substrate specificity to favor viral replication. Few such examples are Coronavirus encoded Papain- like protease (PLpro) and Crimean-Congo hemorrhagic fever virus (CCHFV) encoded L1 protein that has potent de-ubiquitylating and de-ISGylating activity and Human herpesvirus encoded cysteine-proteases (HCMV-UL48 and KSHV-ORF64) that has potent de-ubiquitylating activity. We will identify substrates of these viral proteins involved in the regulation of innate immune response by affinity purification and mass-spectrometry and further elucidate the mode of action and underlying mechanism using molecular-biology, biochemical, gene-knockout/knockdown and imaging methods.
Team leader
Soham Gupta
Assistant Senior LecturerSefanit Rezene
Phd StudentI started my Ph.D. studies in the Systems Virology Group in September 2023. My project focuses on the regulation of host immune responses during viral infections and how viruses may modulate antiviral responses using virus-encoded deubiquitinases (DUBs) and deISGylases (DISGs). The ubiquitin system is fundamental in regulating innate immune signaling, and several viruses have been found to encode proteins that can interfere with these responses by deconjugation of ubiquitin and ubiquitin-like polypeptides, like ISG15. My overall aim is to gain mechanistic insight into different viral DUBs and DISGs using a combination of multi-omics approaches with biochemical, immunological, and molecular techniques.
Publications
Cell-type-resolved quantitative proteomics map of interferon response against SARS-CoV-2.
Saccon E, Chen X, Mikaeloff F, Rodriguez JE, Szekely L, Vinhas BS, Krishnan S, Byrareddy SN, Frisan T, Végvári Á, Mirazimi A, Neogi U, Gupta S
iScience 2021 May;24(5):102420
Type-I interferon signatures in SARS-CoV-2 infected Huh7 cells.
Chen X, Saccon E, Appelberg KS, Mikaeloff F, Rodriguez JE, Vinhas BS, Frisan T, Végvári Á, Mirazimi A, Neogi U, Gupta S
Cell Death Discov 2021 May;7(1):114
