Maria Isaguliants (Issagouliantis) Project Group
Our field of research is centered on genetic or “naked” DNA vaccines.
Maria Issagouliantis
PhD, Docent in ImmunologyResearch
What is a genetic vaccine? Basically, it is a microbial gene (or gene composition) which, when expressed in the cells of vaccine recipient, induce an immune response preventing viral infection and/or exterminating already infected cells. Gene immunogen is composed of the polynucleotide sequence encoding microbial antigen, a strong constitutive promoter and a polyadenylation signal which regulate gene expression in the host cells. Genes can be carried by plasmids, recombinant viruses or bacterial vectors. First genetic vaccines employed natural pathogen-derived coding sequences. They were poorly expressed, their performance was affected by the adverse properties of the microbial proteins (often toxic and/or immunosuppressive). The break-through came with the development of genetic engineering. It allowed to make cardinal changes of the microbial antigens: fuse them to each other, delete toxic/harmful domains, shuffle domains to devoid proteins of their harmful properties while preserving the epitopes, even insert additional domains enhancing the immunogenicity.
For variable microbes it opened a possibility to design consensus proteins represented clusters of closely related/homologous, but still different antigens. Amino acid consensuses built on multiple alignments of carefully selected microbial protein sequences are translated back into genes, respective genes are synthesized with the codons adapted to the organism in which the vaccine would be used. Codon optimization (“humanization” for adaptation to human cells) is done to increase the level of immunogen expression in the host cells and thus increase the immunogenicity. Codon-optimized gene immunogens support >10-fold higher levels of antigen expression as compared to the parental microbial genes. Synthetic genes are cloned into gene immunization vectors to yield naked DNA immunogens. Such expression-optimized consensus gene immunogens are much more efficient than both the parental microbial genes, and their expression-optimized replicas, for example, they can efficiently prevent experimental infection of mice with a lethal dose of influenza virus.
Limiting factors in gene immunization/vaccination in large specifies, as humans, is gene vaccine delivery. First gene vaccines were administered by intramuscular or intradermal injections and resulting levels of in vivo transfection were low. Huge advance was made by delivering DNA by needle-free devices (as biojectors propelling vaccine under high pressure of inert gas), or by electroporation via creating electromagnetic field which open cell pores and helps the injected vaccine to enter cells, or by combination of two methods. Such delivery significantly increases the rate of in vivo transfection, i.e. the number of cells expressing gene immunogen, and, hence, the amount of antigen formed and the strength of the resultant immune response.
Our DNA vaccine project develops candidate vaccines for immunization against chronic viral infections, as HIV-1 and HCV, intended to provide immune pressure on the virus to help the body to control virus replication and in perspective also adding the effects of antiviral drugs.
For human hepatitis C virus, target viral antigens are core protein, nonstructural protein NS5A and RNA-dependent RNA-polymerase NS5B. For HIV-1, the target viral antigens are viral enzymes crucial for viral replication. We hypothesize that immunization with these genes prior to or in parallel to antiviral therapy may prevent the development of resistance to antiviral drugs. The aim of the project is to characterize on the level of cell lines, the properties of these proteins which define their immunogenicity in animal models and ultimately in humans, and develop methods to reduce their potentially harmful effects and increase immunogenicity, i.e. turn them into potent gene vaccine candidates.
Students are welcomed in to participate in design and testing of gene vaccine candidates, also by applying the fascinating technique of in vivo imaging.
Optimization of viral immunogens
First step is the optimization of genes encoding viral antigens. In this project, we elaborate new and improve the existing approaches to viral gene optimization including truncation, deletion of harmful domains & signatures, domain shuffling, insertion of exogenous signals of intracellular antigen retargeting and processing. These studies were started by Elizavet Starodubova on a model antigen - weakly immunogenic reverse transcriptase (RT) of HIV-1. RT was targeted to proteasomal processing by fusing to ornithine decarboxylase (Starodubova E et al, 2008), to lysosomal processing, by fusing to lysosome associated membrane protein 1 (Starodubova E et al, 2010) and to Gly-Ala repeat of EBNA 1 protein of EBV (Starodubova E et al, 2012) (Fig. 1)
The latter two re-routing strategies led to a significant enhancement of RT gene immunogenic performance (Starodubova E et al, 2010; Starodubova E at al, 2012). Immunogenic performance of RT gene was further improved by codon-optimization (Isaguliants M et al, Human Vaccines & Immunotherapeuticals 2013). Immunogenicity tests in mice of the panel of RT gene variants led to elucidation of the driving mechanism behind RT immunogenicity, namely induction of the oxidative stress and oxidative stress response.
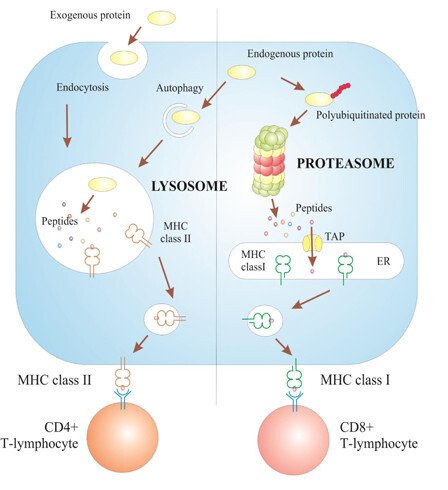
Along these lines, Olga Krotova and Elizaveta Starodubova developed series of HIV gene immunogens targeting HIV-1 integrase (IN). Consensus wild-type and drug-resistant of HIV-1 clade A strain FSU-A prevalent on the territory of the former Soviet Union were designed, and efficiently expressed in pro- and eukaryotic cells. Consensus integrase was shown to be more enzymatically active than enzymes derived from individual viral isolates (Krotova O et al, Biochimie 2013). Introduction of primary drug resistance mutations signatures into integrase backbone yielded, as expected, integrase variants highly resistant to integrase inhibitors raltegravir and elvitegravir (Krotova O et al, Biochimie 2013). The data represent the first study describing the enzymatic properties of the consensus integrase of HIV-1 clade A and the effects of the resistance mutations relieved from the complex effects of sporadic sequence polymorphisms. All HIV integrase variants were shown to be highly immunogenic in mice, inducing polyfunctional Th1-type CD4+ and CD8+ T cells (Krotova OA et al, PLoS One 2013). Generation of such response is highly desirable for an effective HIV-1 vaccine as it offers a possibility to attack virus-infected cells via both MHC class I and II pathways. Another related project led by Athina Kilpeläinen deals with the design and testing of another family of consensus DNA immunogen targeting HIV protease (see Athinas project project description below).
We develop also gene immunogens for HCV core and RNA-dependent RNA-polymerase, where we apply same principle of antigen modification, consensus design and gene optimization. Hepatitis C related gene vaccine experiments are done in tight collaboration with Dr Irina Sominskaya, Biomedical research and Study Center, Riga, Latvia, and Gamaleja Center of Epidemiology and Microbiology, Moscow, Russia, in the frame of the Thematic Partnership of the Swedish Institute “Baltic Network against life-threatening viral infections”
Students are readily accepted for short and long term research projects linked to gene vaccine design and experimental testing.
In vivo methods of monitoring DNA immunization efficacy
Gene vaccine development is impossible without adequate methods to monitor immunogen delivery and expression. Until now that was achieved ex vivo, by analyzing gene expression in biopsies collected after gene immunization. This does not permit to connect gene delivery with the consequent development of immune response. To make that link we suggested to use the in vivo imaging technique. For this, DNA immunogens are administered together with genes encoding bioluminescent reporters, such as the luciferases (Fig. 2). Luciferase activity at the injection site reflects the quality of gene vaccine delivery and initial expression. Activity is seen as bioluminescence is recorded by a Charged Coupled Device (CCD) camera with subsequent conversion of photons that strike a CCD pixel into spatially electric charges (In Vivo Imaging System, IVIS).
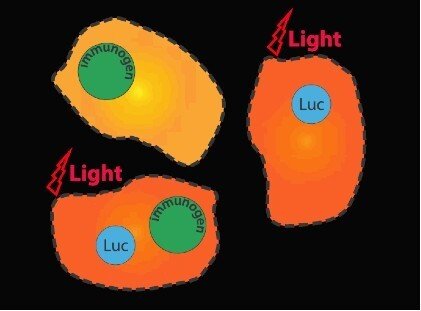
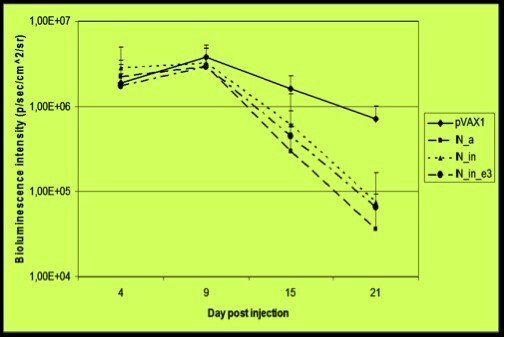
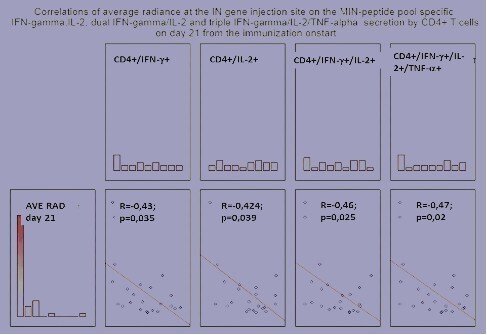
Use of IVIS for optimization of gene immunization constitutes the essence of the research project of Stefan Petkov initiated in 2012. Stefan shown IVIS to be a powerful instrument to optimize the delivery technique (Petkov S et al, 2013). Furthermore, together we found that IVIS can also be used to in vivo monitor the development of immune response. Of two plasmids in immunogen/reporter mixture, only one which encodes gene immunogen can trigger an immune response, the other is “passive” since luciferase is not immunogenic. Development of T cell immune response against gene immunogen correlates with significant decrease of Luc reporter expression. Immune response exterminates reporter and immunogen expressing cells, ultimately leading to the loss of luminescence. In case of immunization with HIV integrase genes, loss of in vivo activity of the luciferase reporter gene co-delivered with plasmids encoding integrase variants correlated with the development of specific multi-cytokine responses of CD8+ and CD4+ T-cells (Fig. 3A, B; Krotova OA et al, PLoS One, 2013). The decrease in reporter activity can be used to quantitatively characterize gene immunogen performance, specifically immunogenicity on T cell level. This allows to select portent gene vaccine candidates on the very early stages of preclinical testing and by this, to significantly reduce and refine animal experimentation.
Currently, we are identifying which T cell (sub)populations are responsible for the decrease of luciferase reporter activity. Using this knowledge will plan to design in vivo imaging alternatives of immune tests assessing activities of specific T cell subsets. Furthermore, we aim to specify the links between the parameters of reporter activity loss (magnitude, kinetics) and types of immune response (innate, cellular of Th1 or Th2 type, antibody) to set a panel of noninvasive in vivo alternatives for immune assays which otherwise request sacrification of small animals.
The project has been made possible thanks to the generous financial support of the program for the Refinement, Reduction and Replacement (3R) of animal experiments of the Swedish Research Council.
Students are readily accepted for short and long term research projects linked to gene vaccine design and experimental testing.
Molecular mechanisms of liver disease induced by hepatitis C virus
Chronic infection with Hepatitis C virus induces liver inflammation, apoptosis, and oxidative stress contributing to multiple metabolic disorders. When untreated, chronic hepatitis C progresses to hepatic decompensation, cirrhosis and hepatocellular carcinoma. Virus has developed a sophisticated strategy to adapt to the host, and to adapt host to the needs of viral replication.
This project dwells with molecular mechanisms underlying viral adaptation strategy and viral pathogenicity.
One arm of the study is centered on HCV polymerase (RdRp or NS5B), its polymorphisms and enzymatic activities which determine HCV replication fitness. In collaboration with University of Dentistry and Medicine, and Ivanovsky Institute of Virology, Moscow, we obtained a panel of HCV polymerases from HCV genotype 1b residing in chronic hepatitis C patients with abnormal and persistently normal liver enzyme levels. RdRp variants are cloned for eukaryotic expression, currently we are setting up reporter assays to assess their activity in expressing cells, to further compare it in between RdRp variants and rely to differences in their amino acid structure. Currently, the project is carried in collaboration with prof John Tavis, St Louis School of Medicine, St Louis, USA and Dr Alex Ivanov, WA Engelhard Institute of Molecular Biology, Moscow, Russia.
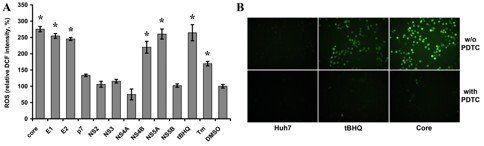
Second arm focuses on the mechanisms of HCV-induced oxidative stress. We have demonstrated that expression in liver cells of HCV nucleocapsid (core) protein leads to induction of strong oxidative stress, and a strong up-regulation of antioxidant defense system. Other HCV proteins, as E1, E2, NS4B, and NS5A also induce ROS production, albeit in a lesser extent (Fig. 4). All five activate antioxidant defense Nrf2/ARE pathway via several independent mechanisms including Nrf2 activation by protein kinase C mediated by ROS, and also by other ROS-independent pathway(s) (Ivanov A et al, 2011). In the current project, we aim to identify the domains of HCV core responsible in ROS induction and NRf2 activation. Altogether, these molecular events may explain the mechanism(s) of oxidative stress during acute stage of hepatitis C. The study is supported by the New Visby program of the Swedish Institute, and Russian Foundation for Basic Research.
Also, jointly with the researchers from the University of Eastern Finland (Kuopio, Finland) and WA Engelhard Institute of Molecular Biology (Moscow, Russia) we are investigating the relationship between HCV replication, oxidative stress and polyamine metabolism. In a recent study, we had demonstrated that oxidative stress in hepatoma cells leads to an enhanced polyamine metabolism as reflected by both increased levels of polyamine biosynthesis by ornithine decarboxylase, and of polyamine degradation by (spermidine/spermine-N(1)-acetyltransferase (Smirnova OA, Isaguliants M, 2012). The balance in these antagonistic activities was shifted towards polyamine biosynthesis, which resulted in increased intracellular levels of putrescine, spermidine, and spermine, favoring cell proliferation. This set-up promotes survival of cells sub-lethally damaged by oxidative stress, and, thus, promotes malignant transformation. We are now testing if such shift towards polyamine biosynthesis occurs also in cells stressed by transient expression of HCV proteins. This research would help to explain the survival of HCV-infected cells, and define one of the molecular mechanisms supporting its persistence and carcinogenicity. The study is supported by the Swedish Institute New Visby program, the University of Eastern Finland, the Russian Ministry of Education and Science and Russian Foundation for Basic Research.
Students are welcomed to join in to perform the research abroad at the collaborating institutions.
Grants
We acknowledge project support from the Swedish Research Council in 2009-2011 and 2011-2014 project nr 2010-3783 (grants to M. Isaguliants), Russian Fund for Basic Research 2011-2013 (grant to E. Starodubova), Russian Fund for Basic Research 2013-2015 (grant to M. Isaguliants 13-04-0152313), and the Thematic Partnership of the Swedish Institute 2013-2016 (project nr 09272_2013).
Microbial vaccines as targeted cargo, genetic approach
Swedish Research Council K2009-66X-21053-01-3; January 1, 2009 – December 31, 2011
Reducing animal experimentation in vaccine development by the use of in vivo imaging techniqueSwedish Research Council K2010-3783; January 1, 2010 – December 31, 2015
Thematic Partnership of the Swedish Institute “Baltic Network against Life-threatening viral infections 2013-2016” (project nr 09272_2013).
Swedish Institute Dnr 09272_2013
Network Coordinator: Maria Issagouliantis, Department of Microbiology, Tumor and Cell Biology, Karolinska Institutet.
New Visby Network on Hepatitis C 2011-2012. Identification, control and prevention of human hepatitis C virus infection
Swedish Institute Dnr 00885/2011; 2011-2013
Network Coordinator: Maria Issagouliantis, Department of Microbiology, Tumor and Cell Biology, Karolinska Institutet.
New Visby Network on Hepatitis C 2010-2011. Identification, control and prevention of human hepatitis C virus infection
Swedish Institute Dnr 00747/2010; 2010-2012
Network Coordinator: Prof Anders Widell, Malmö University Hospital, Lund University, Malmö, Sweden.
Project Group Members
Elizaveta Starodubova Affiliated researcher
WA Engelhard Institute of Molecular Biology, Moscow,
Phone: 08-52485993 E-mail: estarodubova@gmail.com
Elizaveta leads the research project on targeting of DNA immunogens to MHC class II route of processing and presentation. The project is focused on the principles of DNA vaccine design, specifically modulation of immunogenicity of antigens by changing its processing pathway, specifically by retargeting to lysosome using as a model HIV-1 reverse transcriptase of HIV-1.
Stefan Petkov PhD student
Stefan Petkov Research project (Pdf file)
Stefans research project deals with In vivo imaging in application to preclinical trials of genetic vaccines. In vivo optical imaging of bioluminescence has a wide variety of uses including tracking of in vivo gene expression in various tissues, detection of protein–protein interactions, monitoring of tumor growth, etc. The capacity of bioluminescent imaging (BLI) to detect in vivo gene expression through the activity of luminescent enzymes such as luciferases has made it an attractive way of monitoring gene delivery and initial expression by detecting bioluminescence. Project has been made possible thanks to the financial support of the Swedish Research Council.
Athina Kilpelainen Researcher
E-mail: athina.kilpelainen@stud.ki.se
Project of Athina Kilpeläinen deals with the design and testing of another family of consensus DNA immunogen targeting HIV protease. First proof was obtained in mice that plasmid encoding protease mutations of resistance to ritonavir can induce T-cell immune response recognizing individual mutations. Results reported on 11th meeting of Baltic Network against life-threatening viral infections, and published as abstracts in Medical Virology ISSN 2070-7746 vol XXVIII, nr 4; abstracts available on the Network web-site at http://mtcexternal.ki.se.preview.binero.se/Baltic-Antiviral-Network/, and on the 4th Conference “Vaccines and Vaccinations” Valencia, Spain, 2014 (Isagulaints M et al, J Vaccines and Vaccination, 2014).
Sviataslau Sasinovich Stipendiate
PhD student
E-mail: sviataslau.s@gmail.com
The aim of this project is to characterize on the molecular level HIV and HCV epidemics in Belarus. On the materials collected in his previous studies, Sviat plans to characterize: (1) HIV and HCV viral strains circulating in Belarus and trace their origins and direction of evolution; (2) HIV-1 mutations /patterns of mutations conferring resistance to standard and newly introduced antiretroviral drugs; (3) HCV mutations/mutation patterns conferring resistance to directly acting antiviral drugs; (4) frequency and routes of primary infections with drug resistant HIV and HCV. The project is carried in collaboration with Prof Patrik Medstrand, Department of Laboratory Medicine in Malmö Lund University.
Kevin Fallahi, student
Karolinska Institutet
kevin.fallahi@stud.ki.se
Tuomo A. Keinänen, Guest researcher
2013-2014, Associate Professor in chemical biology , University of East Finland, Kuopio, Finland; tuomo.keinanen@uef.fi
Olga Krotova, PhD student & guest researcher
2012-2014; Gamaleja Research Center for Epidemiology and Microbiology, Moscow, Russia, oakrotova@inbox.ru
Research collaborations
- Prof Anders Widell, Malmö University Hospital, Lund University, Malmö, Sweden;
- Prof Patrik Medstrand, Department of Laboratory Medicine in Malmö Lund University.
- Prof Jorma Hinkula, University of Linköping, Linköping, Sweden;
- Dr Birke Bartosch, INSERM, Lyon, France;
- Prof John Tavis, Department of Molecular Microbiology and Immunology, St Louis University School of Medicine, St Louis, USA;
- Dr Jan Drobeniuc, Centers for Disease Control and Prevention (CDC), Atlanta, USA;
- Dr Saleem Karimi, Centers for Disease Control and Prevention (CDC), Atlanta, USA;
- Prof. Mauro Magnani, Centre of Biotechnology, Urbino University, and DIATHEVA s.r.l., Italy;
- Prof Yves Rivier, Pasteur Institute, Paris, France; Prof Vladimir Lukashov, University of Amsterdam, the Holland;
- Dr Irina Sominskaya, Biomedical Research and Study Center, Riga, Latvia;
- Dr Valentina Tefanova, National Institute for Health Development, Tallinn, Estonia;
- Dr Saulius Caplinskas, Center for Communicable Diseases and AIDS, Vilnius, Lithuania;
- Dr Jin-Ching Lee, College of Life Science, Kaohsiung Medical University, Kaohsiung City, Taiwan;
- Dr Santiago Dueñas-Carrera, Centro de Ingeniería Genética y Biotecnología, Havana, Cuba;
- Dr Alexander Ivanov, Engelhardt Institute of Molecular Biology, Moscow;
- Dr Olga Znoyko, Moscow State University of Dentistry and Medicine, Moscow, Russia;
- Prof Marina Gottikh, Lomonosov Moscow State University, Moscow, Russia;
- Prof Sergey Kochetkov, Engelhardt Institute of Molecular Biology, Moscow;
- Prof Vadim Karpov, Engelhardt Institute of Molecular Biology, Moscow;
- Dr Olga Kalinina, Pasteur Institute, St Petersburg, Russia.
Selected Publications
A qPCR assay for measuring the post-integrational DNA repair in HIV-1 replication.
Anisenko AN, Knyazhanskaya ES, Isaguliants MG, Gottikh MB
J Virol Methods 2018 12;262():12-19
DNA immunization site determines the level of gene expression and the magnitude, but not the type of the induced immune response.
Petkov S, Starodubova E, Latanova A, Kilpeläinen A, Latyshev O, Svirskis S, et al
PLoS One 2018 ;13(6):e0197902
Codon optimization and improved delivery/immunization regimen enhance the immune response against wild-type and drug-resistant HIV-1 reverse transcriptase, preserving its Th2-polarity.
Latanova AA, Petkov S, Kilpelainen A, Jansons J, Latyshev OE, Kuzmenko YV, et al
Sci Rep 2018 05;8(1):8078
Consensus HIV-1 subtype A integrase and its raltegravir-resistant variants: design and characterization of the enzymatic properties.
Shadrina O, Krotova O, Agapkina J, Knyazhanskaya E, Korolev S, Starodubova E, et al
Biochimie 2014 Jul;102():92-101
Evaluation of immunogen delivery by DNA immunization using non-invasive bioluminescence imaging.
Petkov SP, Heuts F, Krotova OA, Kilpelainen A, Engström G, Starodubova ES, et al
Hum Vaccin Immunother 2013 Oct;9(10):2228-36
Oxidative stress induced by HIV-1 reverse transcriptase modulates the enzyme's performance in gene immunization.
Isaguliants M, Smirnova O, Ivanov AV, Kilpelainen A, Kuzmenko Y, Petkov S, et al
Hum Vaccin Immunother 2013 Oct;9(10):2111-9
Consensus HIV-1 FSU-A integrase gene variants electroporated into mice induce polyfunctional antigen-specific CD4+ and CD8+ T cells.
Krotova O, Starodubova E, Petkov S, Kostic L, Agapkina J, Hallengärd D, et al
PLoS One 2013 ;8(5):e62720
Cellular immunogenicity of novel gene immunogens in mice monitored by in vivo imaging.
Starodubova E, Krotova O, Hallengärd D, Kuzmenko Y, Engström G, Legzdina D, et al
Mol Imaging ;11(6):471-86
Chemically induced oxidative stress increases polyamine levels by activating the transcription of ornithine decarboxylase and spermidine/spermine-N1-acetyltransferase in human hepatoma HUH7 cells.
Smirnova OA, Isaguliants MG, Hyvonen MT, Keinanen TA, Tunitskaya VL, Vepsalainen J, et al
Biochimie 2012 Sep;94(9):1876-83
A combination of intradermal jet-injection and electroporation overcomes in vivo dose restriction of DNA vaccines.
Hallengärd D, Bråve A, Isaguliants M, Blomberg P, Enger J, Stout R, et al
Genet Vaccines Ther 2012 Aug;10(1):5
Hepatitis C virus proteins activate NRF2/ARE pathway by distinct ROS-dependent and independent mechanisms in HUH7 cells.
Ivanov AV, Smirnova OA, Ivanova ON, Masalova OV, Kochetkov SN, Isaguliants MG
PLoS One 2011 ;6(9):e24957
Increased expression and immunogenicity of HIV-1 protease following inactivation of the enzymatic activity.
Hallengärd D, Haller BK, Petersson S, Boberg A, Maltais AK, Isaguliants M, et al
Vaccine 2011 Jan;29(4):839-48
The successful immune response against hepatitis C nonstructural protein 5A (NS5A) requires heterologous DNA/protein immunization.
Masalova OV, Lesnova EI, Pichugin AV, Melnikova TM, Grabovetsky VV, Petrakova NV, et al
Vaccine 2010 Feb;28(8):1987-96
Recent Reviews
DNA vaccination against drug resistance in chronic viral infections. Proceedings of 4th International Conference on Vaccines and Vaccination. J Vaccine and Vaccination, 2014, 5:5
Regulation of Immunogen Processing: Signal Sequences and Their Application for the New Generation of DNA-Vaccines.
Starodubova ES, Isaguliants MG, Karpov VL
Acta Naturae 2010 Apr;2(1):53-60
Vaccination against drug resistance in HIV infection.
Boberg A, Isaguliants M
Expert Rev Vaccines 2008 Feb;7(1):131-45
Functionality of the immune system in patients with chronic hepatitis C: trial by superinfections and vaccinations.
Isaguliants MG
Expert Rev Vaccines 2007 Aug;6(4):527-37
