Research theme: Biotechnology
After more than two decades of efforts, we subsequently succeeded to develop production in E. coli of recombinant selenoproteins having internal Sec residues, which had not been possible earlier. This was enabled using UAG (instead of UGA) as the codon for Sec in an RF1-deficient host strain, and represented a major methodological breakthrough, as it opened recombinant production for virtually any selenoprotein. The methodology gives us a unique cutting-edge advantage in studies of selenoproteins and today provides a very important methodological foundation for our research.
Recombinant selenoprotein production in E. coli
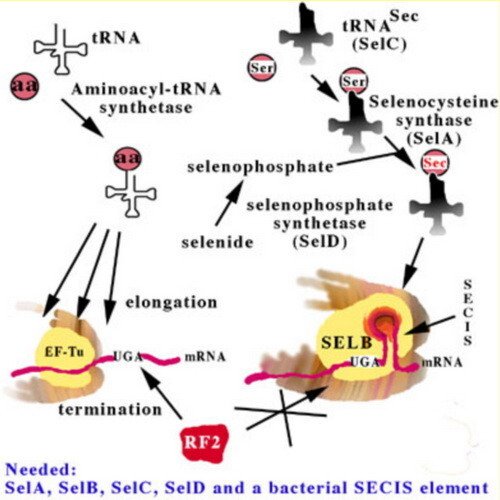
In order to comprehend heterologous selenoprotein synthesis in bacteria, it is first necessary to review the main aspects of the bacterial selenoprotein translation machinery. This machinery is highly complex, as has been revealed in great detail by August Böck and coworkers. The critical elements are summarized in this figure.
The selenocysteine-specific elongation factor, SELB, binds to a secondary structure in the mRNA, a Selenocysteine Insertion Sequence (SECIS) element, following the UGA codon encoding Sec insertion (instead of termination by release factor 2 as encoded by UGA in non-selenoprotein genes). SELB catalyzes Sec insertion using a selenocysteinylated selenocysteine-specific tRNA, the SelC gene product, which originally is charged with a seryl residue, but is converted to a selenocysteinyl moiety by the SelA gene product, selenocysteine synthase, using selenophosphate as selenium donor, formed from selenide by selenophosphate synthetase, the SelD gene product. All in all, is the factors needed for selenoprotein synthesis in E. coli are the four SelA - D gene products and a SELB-compatible SECIS structure in the mRNA.
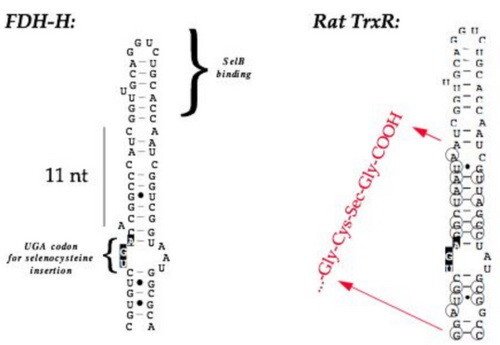
Mammalian selenoprotein mRNA's also contain a SECIS element, but with other structural characteristics and they are positioned in the 3'-UTR. This is not compatible with the E. coli translation machinery - for that reason mammalian selenoproteins are not possible to directly express in E. coli, but will always lead to termination of translation at the UGA selenocysteine codon. However, by tailoring a selenoprotein gene to contain a bacterial-type SECIS element, this makes it possible to express the product in E. coli.
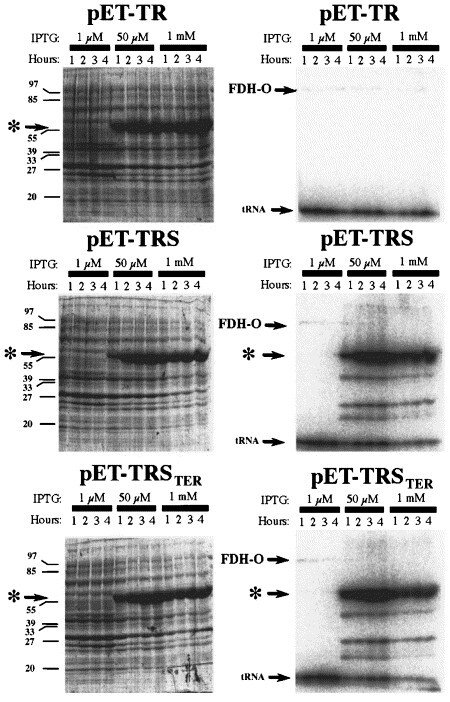
By engineering an E. coli-type SECIS element in the gene for rat thioredoxin reductase 1, it became possible to express the protein as a selenoprotein recombinantly in the bacteria, which is illustrated in this figure. On the left-hand side are Commassie-stained gels whereas the right-hand figures are autoradiograms of the same gels, showing 75Se-selenite labelled products. "pET-TR" encoded TrxR1 without a SECIS element in the gene, pET-TRS contained a natural formate dehydrogenase-derived SECIS element, whereas pET-TRSTER contained a SECIS element encoding the TrxR1 carboxyterminus (see first figure, above). Note that the presence of a SECIS element was necessary for selenium incorporation, but that when present the reserve capacity of E. coli to synthesize selenoproteins is widely surpassing the normal synthesis under aerobic growth of the natural FDH-O selenoprotein (indicated in the figure).
The expression of rat TrxR1 was described in "High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes".
The method of heterologous selenoprotein production is further discussed in the review "Recombinant expression of mammalian selenocysteine-containing thioredoxin reductase and other selenoproteins in Escherichia coli".
It was further optimized in the study "Assessment of production conditions for efficient use of Escherichia coli in high-yield heterologous recombinant selenoprotein synthesis".
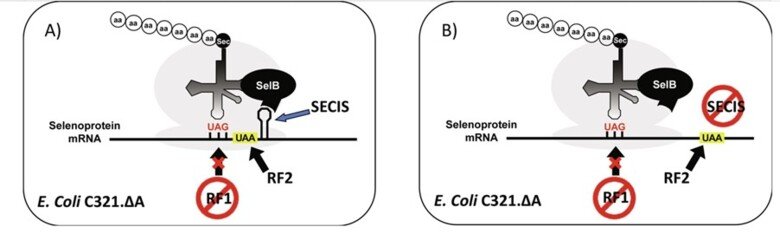
For further information, see "Selenocysteine Insertion at a Predefined UAG Codon in a Release Factor 1 (RF1)-depleted Escherichia coli Host Strain Bypasses Species Barriers in Recombinant Selenoprotein Translation", "Production and purification of homogenous recombinant human selenoproteins reveals a unique codon skipping event in E. coli and GPX4-specific affinity to bromosulfophthalein" and "Expressing recombinant selenoproteins using redefinition of a single UAG codon in an RF1-depleted E. coli host strain".
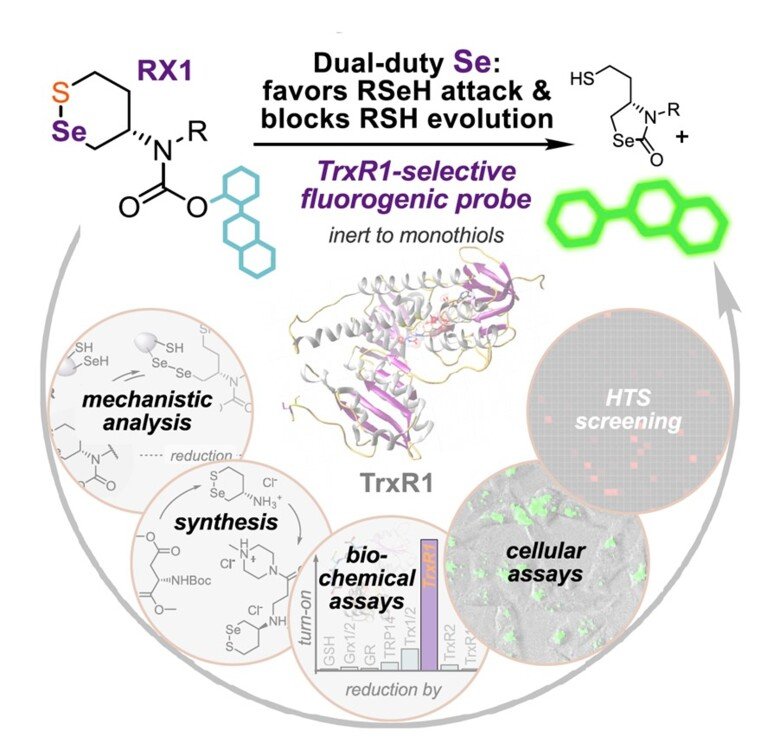
RX1 – a highly specific fluorescent probe for TrxR1 activity
Together with Oliver Thorn-Seshold in Munich, Germany, we found that previously claimed “TrxR1-specific” probes for its activity were actually highly promiscuous and should not be used for assessment of TrxR1 activity "Cyclic 5-membered disulfides are not selective substrates of thioredoxin reductase, but are opened nonspecifically". However, using a completely novel molecular scaffold with a selenenylsulfide motif it was possible to develop a truly specific probe for detection of TrxR1 activity by fluorescence, which we hvae validated extensively using pure enzyme systems as well as in cells ("Selective cellular probes for mammalian thioredoxin reductase TrxR1: rational design of RX1, a modular 1,2-thiaselenane redox probe"). This unique probe for specific assessment of TrxR1 activity in cells provides and excellent tool for in-depth studies of the activities of this selenoprotein within a cellular context.
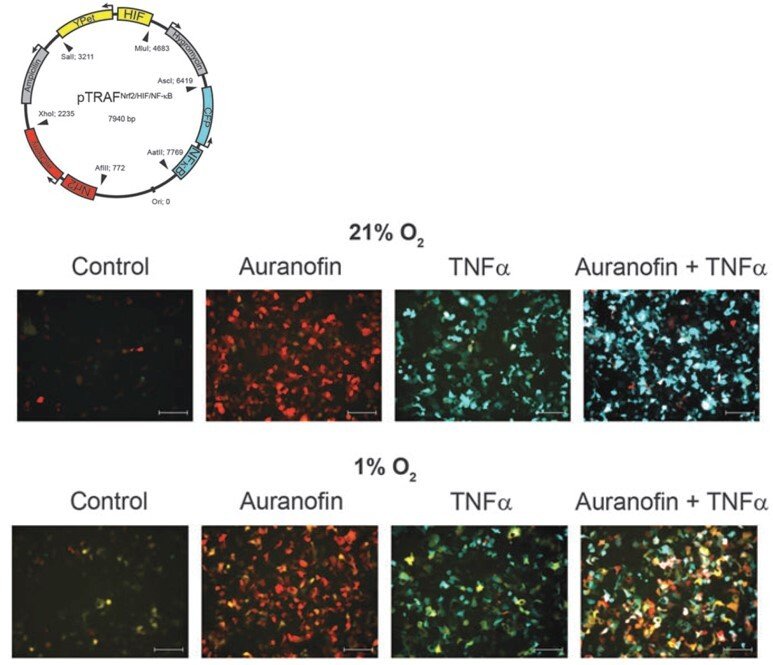
pTRAF – probing several transcription factor activation patterns at single-cell resolution
Wishing to understand how different transcription factors can be separately regulated by redox control within the same cellular context, we developed a pTRAF tool (plasmid for transcription factor reporter activation based on fluorescence) that enables simultaneous determination of Nrf2, HIF and NFκB activation patterns at single-cell resolution. The performance of the pTRAF was validated using cell cultures and spheroids, showing how redox perturbing drugs and stem cells directly regulate these transcription factors ("Time- and cell-resolved dynamics of redox-sensitive Nrf2, HIF and NF-κB activities in 3D spheroids enriched for cancer stem cells" and "Cross Talk in HEK293 Cells Between Nrf2, HIF, and NF-κB Activities upon Challenges with Redox Therapeutics Characterized with Single-Cell Resolution"). The pTRAF represents another useful tool for the assessment of redox regulation.
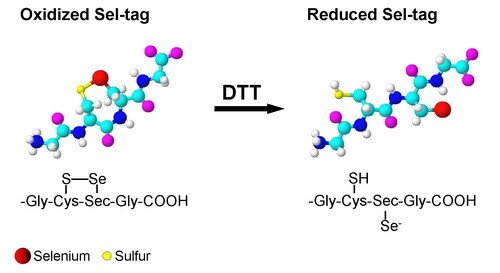
Development of a Sel-tag
Inspired by the properties of the C-terminal tetrapeptide motif of mammalian TrxR, we have developed a redox active Sel-tag for use with recombinant proteins. We found that this Sel-tag could be used for one-step purification of the tagged protein, selenolate-targeted fluorescent labeling, as well as selenolate-tageted radiolabeling, with either selenium-75 or with the positron-emitter carbon-11. Further development of this technique is currently a highly prioritized issue in our laboratory.
The results were published in the launch issue of the new Nature Methods journal and titled "Exploiting the 21st amino acid-purifying and labeling proteins by selenolate targeting".
A detailed protocol of utilizing this technique was published in the Nature Protocols journal and titled "Tagging recombinant proteins with a Sel-tag for purification, labeling with electrophilic compounds or radiolabeling with 11C".
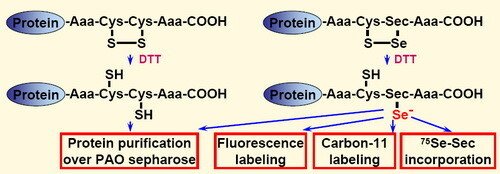
To explore the importance of selenocysteine in such applications, we here analyzed four different redox active C-terminal motifs, carrying either dithiol (Gly-Cys-Cys-Gly or Ser-Cys-Cys-Ser) or selenolthiol (Gly-Cys-Sec-Gly or Ser-Cys-Sec-Ser) motifs. Utilizing these different functional motifs with the same recombinant protein (Fel d 1), we could assess their relative reactivity and potential usefulness for biotechnological applications. Either dithiol or selenolthiol tetrapeptide tags on recombinant proteins can be efficiently used in one-step purification via PAO sepharose. In addition, the selenolthiol variants (but not the dithiol tags) were highly reactive in rapid labeling with electrophilic fluorescent or the short-lived positron emitting compound 11CH3I. As a unique feature for selenocysteine-containing proteins, the selenolthiol variants also permit methodologically simple in situ [75Se]-g-emitting radiolabeling through recombinant [75Se]-Selenocysteine incorporation.
This work was described in "Selenolthiol and dithiol C-terminal tetrapeptide motifs for one-step purification and labeling of recombinant proteins produced in E. coli".
