Medical Digital Twin Research Group
One of the most important health care problems is that many patients with common disease like immunological, malignant and cardiovascular diseases do not respond to treatment. This reflects the complexity and heterogeneity of diseases. Each disease may involve thousands of genes, which vary across billions of cells in many organs, as well as between time points and patients with the same diagnosis. There is a wide gap between this complexity and modern health care.
We aim to address this gap by digital twins (DTs) of individual patients, as recently discussed by us in Nature Biotechnology.
DTs are computational models based on clinical routine and multi-omics data, down to the single cell level. DTs are computationally treated with thousands of drugs to find the optimal drug or drug combination for individual patients (see figure below).
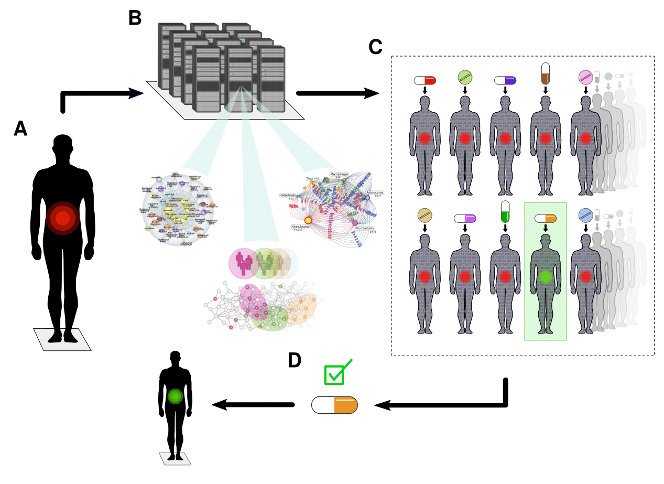
Our long-term aim is that each willing individual will be offered her/his own DT for early prediction and prevention of disease. Our group consists of clinical, bioinformatics, omics and experimental researchers, which are also part of the Swedish Digital Twin Consortium.
Visiting address
Widerströmska Huset
Tomtebodavägen 18A
171 65 Solna
Delivery address
Medical Digital Twin Research Group
Tomtebodavägen 18A
171 65 Solna
Learn more?
Want to learn more about us and how researchers can use Digital Twins for predictive, preventive and personalised medicine?
Please visit the SDTC website, which contains links links to an EU initiative aiming at DT for personalised medicine, publications, recorded talks at Harvard and SciLifeLab, publications, TV interviews and a music video created by collaborating artists and a composer at the Royal College of Music.
Group Members
Mikael Benson
Principal ResearcherXinxiu Li
Research SpecialistResearch Specialist, responsible for clinical translation of DTs.
Hui Wang
Affiliated to ResearchVisiting Professor, responsible for experimental validation.
Yelin Zhao
Postdoctoral ResearcherPost doctoral fellow, responsible for DT construction and analyses.
Jonathan Dorairaj
Research AssistantResearch Data Scientist.
Francesca Borsani
Masters student in precision medicineSelected Publications
Martin Smelik, Daniel Diaz-Roncero Gonzalez, Xiaojing An, Rakesh Heer, Lars Henningsohn, Xinxiu Li, Hui Wang, Yelin Zhao, Mikael Benson. Combining spatial transcriptomics, pseudotime and machine learning to find biomarkers for prostate cancer. Cancer Research 2025;85(13):2514-2526. doi: 10.1158/0008-5472.CAN-25-0269.
Li X, Loscalzo J, Mahmud AKMF, Aly DM, Rzhetsky A, Zitnik M, Benson M. Digital twins as global learning health and disease models for preventive and personalized medicine.Genome Med. 2025 Feb 7;17(1):11. doi: 10.1186/s13073-025-01435-7
Schäfer S, Smelik M, Sysoev O, Zhao Y, Eklund D, Lilja S, Gustafsson M, Heyn H, Julia A, Kovács IA, Loscalzo J, Marsal S, Zhang H, Li X, Gawel D, Wang H, Benson M. scDrugPrio: A framework for the analysis of single-cell transcriptomics to address multiple problems in precision medicine in immune-mediated inflammatory diseases. Genome Med 2024,
Lilja S, Li X, Lee EJ, Loscalzo J, Zhang H, Zhao Y, Gawel D, Wang H, Benson M. Multi-organ single cell analysis reveals an on/off switch system with potential for personalized treatment of immunological diseases. Cell Reports Medicine 2023:(3):100956. doi: 10.1016/j.xcrm.2023.100956
Li X, Lee EJ, Lilja S, Bohle B, Loscalzo J, Schäfer S, Zhang H, Gustafsson M, Zhao Y, Gawel D, Wang H, Benson M. A dynamic single cell-based framework for digital twins to prioritize disease genes and drug targets. Genome Medicine 2022
Mansour Aly D, Dwivedi OP, Prasad RB, Käräjämäki A, Hjort R, Thangam M, Åkerlund M, Mahajan A, Udler MS, Florez JC, McCarthy MI; Regeneron Genetics Center, Brosnan J, Melander O, Carlsson S, Hansson O, Tuomi T, Groop L, Ahlqvist E. Genome-wide association analyses highlight etiological differences underlying newly defined sub types of diabetes Nat Genet. 2021 Nov;53(11):1534-1542.
Björnsson B, Borrebaeck C, Elander N, Gasslander T, Gawel DR, Gustafsson M, Jörnsten R, Lee EJ, Li X, Lilja S, Martínez-Enguita D, Matussek A, Sandström P, Schäfer S, Stenmarker M, Sun XF, Sysoev O, Zhang H, Benson M; Swedish Digital Twin Consortium. Digital twins to personalize medicine. Genome Medicine 2020;12(1):4.
Jia G, Li Y, Zhang H, Chattopadhyay I, Boeck Jensen A, Blair DR, Davis L, Robinson PN, Dahlén T, Brunak S, Benson M, Edgren G, Cox NJ, Gao X, Rzhetsky A. Estimating heritability and genetic correlations from large health datasets in the absence of genetic data. Nature Communications. 2019;10(1):5508
Lentini M, Lagerwall M, Vikingsson S, Mjoseng HK, Vogt H, Green H, Meehan RR, Benson M*, Nestor CE*. Pervasive experimental errors have severely distorted our understanding of mammalian epigenetics. Nature Methods 2018; 15:499-50.
Shalek AK, Benson M. From bench to bedside: Single-cell RNA-sequencing to personalize medicine. Science Translational Medicine 2017;9(408)
Mika Gustafsson.. Colm E. Nestor, Mikael Benson. A validated gene regulatory network and GWAS identifies early regulators of T-cell associated diseases. Science Translational Medicine 2015;7(313):313ra178
S. Bruhn, Y. Fang, F.Barrenäs, M. Gustafsson,H. Zhang, A Konstantinell, A. Krönke, B. Sönnichsen, A. Bresnick, N. Dulyaninova, H. Wang, Y. Zhao, J. Klingelhöfer, N Ambartsumian, MK, Beck, C. Nestor, E Bona, Z. Xiang, M. Benson. A generally, applicable module-based translational strategy identifies a key diagnostic and therapeutic candidate gene in allergy. Science Translational Medicine 2014:8;6
