Project: Transcriptional mutagenesis and cellular effects
This research proposes to shift the long-standing emphasis from the effects of mutagenesis from replication past DNA lesions to the role DNA damage plays in RNA synthesis.
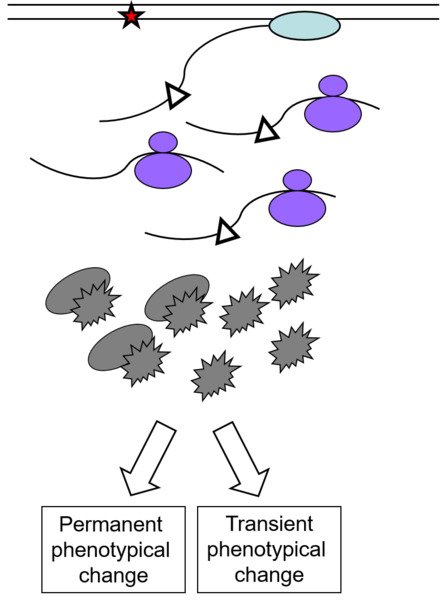
Frequently occurring non-bulky lesions, such as 8-oxoguanine (8oxoG), and O6-methylguanine (O6meG), are efficiently bypassed by RNA polymerases in vitro. This could induce incorporation of incorrect bases into nascent RNA, potentially altering the transcript's function via a process called transcriptional mutagenesis (TM). Recent studies have reported the ability of 8oxoG and O6meG to induce TM in mammalian cells affecting protein functions. However, the relative importance of TM in human health is still virtually untouched.
Although the concept of transcriptional errors (i.e. misincorporations) is well known, the biological consequences are still a recent discovery. Available data show that transcriptional errors can lead to reduced splicing fidelity, production of mutant protein with altered function, reprogramming of cellular signaling and transcriptional networks and shortening of cellular lifespan. While the few existing studies on this topic have begun to shed light on the process, many issues have yet to be addressed. The extent, to which transcriptional errors occurs in mammalian cells and the role these seemingly transient errors might play in disease processes, is currently unknown. Current theories postulate that the transient expression of mutant proteins due to transcriptional errors could play a major role in the development of concomitant changes in cell physiology, potentially triggering pathology. In order to characterize each of these events, an approach based on RNAseq and reporter plasmids has been developed for studying the occurrence and potentially detrimental cellular effects originating from transcriptional errors. Of especial interest is also the impact of DNA damage on the fidelity of transcription (referred to as transcriptional mutagenesis, TM) and its consequences.
The project is performed in collaboration with Dr. Vicent Pelechano's group at SciLife Lab, Karolinska Institutet.
Financing
- Carl Tryggers Foundation
- IMM Junior Faculty
- Seventh framework Marie Curie international reintegration grant (IRG)
- Swedish Research Council (VR)
Publications
Transcriptional mutagenesis dramatically alters genome-wide p53 transactivation landscape.
Liang S, Ezerskyte M, Wang J, Pelechano V, Dreij K
Sci. Reports. 2020 Aug;10:13513
O6-methylguanine-induced transcriptional mutagenesis reduces p53 tumor-suppressor function.
Ezerskyte M, Paredes JA, Malvezzi S, Burns JA, Margison GP, Olsson M, Scicchitano DA, Dreij K
Proc. Natl. Acad. Sci. U.S.A. 2018 05;115(18):4731-4736
Transcriptional mutagenesis reduces splicing fidelity in mammalian cells.
Paredes JA, Ezerskyte M, Bottai M, Dreij K
Nucleic Acids Res. 2017 Jun;45(11):6520-6529
O6-methylguanine induces altered proteins at the level of transcription in human cells.
Burns JA, Dreij K, Cartularo L, Scicchitano DA
Nucleic Acids Res. 2010 Dec;38(22):8178-87
DNA damage and transcription elongation: consequences and RNA Integrity.
Dreij K, Burns J, Dimitri A, Nirenstein L, Noujnykh T, Scicchitano DA
In The Chemical Biology of DNA Damage. N. Geacintov, S. Broyde, eds. Wiley-VCH Verlag, Weinhem. Chapter 17, 399-437. 2010.
