The Small Molecule Mass Spectrometry Core Facility (KI-SMMS) - our offer
KI-SMMS provides an array of sensitive and specific methods to quantify small molecules in all types of sources. Our main goal is the generation of high-quality mass-spectrometry based data that fulfils the specific needs of each user. To that end, we can assist you with all parts of the pipeline, from experimental design to data interpretation.
The Small Molecule Mass Spectrometry Core Facility (KI-SMMS) offers services to analyse small molecules using the most specific and sensitive targeted mass spectrometry-based methods available.
Our services include broad metabolomics and lipidomics screening panels for the relative quantification of these small molecules.
We also provide full quantitation for selected panels including polar metabolites and lipids, as well as for xenobiotics like drugs, vitamins, and their biotransformation products.
Custom methods can also be developed on request.
We have decades-long cumulative experience servicing both academic and industrial projects. Whatever your question is, we make sure that the analyses are tailored to answer it. We focus on helping researchers by conducting metabolomic, lipidomic and other mass spectrometry-based experiments and analyses.
Our activity is focused on:
- Supply researchers with the tools and resources they need to explore and understand biological phenomena at the small molecule level
- Develop, adapt and establish novel approaches that anticipate future needs of researchers
- Transversal technological research with other internal or external teams (platforms, research units)
- Teaching and training users about the basics and possibilities that mass spectrometry offers in small molecule analysis
Let us make you an offer
Not sure what you want, how to do it or how much it will cost? Please contact us, schedule a meeting and/or book our services in iLab.
Our services
A global panel for the characterization of small molecules commonly present in biological matrices. Compounds are characterized using their chromatographic retention times and one selected reaction monitoring (SRM) transition. For most compounds, a second SRM is used for additional confirmation. Typical families of compounds screened include:
- Amino acids and derivatives
- TCA metabolites
- Nucleotides and nucleosides
- Carbohydrates
- Organic acids
- Vitamins and their metabolites
- Bile acids
- Carnitines
Check our panels in our Overview of current panels (excel file) below.
If not present, your metabolite of interest can, if technically feasible, be included on demand in the panel.
A global panel for the characterization of lipid species from the most common families commonly present in biological matrices. Compounds are characterized using their chromatographic retention times and, at least, one class-specific selected reaction monitoring (SRM) transition. Lipids screened belong to the following classes:
- Sphingolipids
- Phosphatidylcholines
- Phosphatidylethanolamines
- Phosphatidylserines
- Diacylglycerols
- Triacylglycerols
- Fatty acids
Check our panels in our Overview of current panels (excel file) below.
If not present, your lipid of interest can, if technically feasible, be included on demand in the panel.
A global panel for the quantification of oxylipins derived from different polyunsaturated fatty acids. We have experience commonly present in biological matrices. Compounds are characterized using their chromatographic retention times and, at least, one class-specific selected reaction monitoring (SRM) transition.
Different panels exist, depending on the oxylipins and matrix of interest.
Check our panels in our Overview of current panels (excel file) below.
Custom designed experiments to follow the metabolic reactions of your compounds.
Includes method development and sample analyses for the detection and quantitation of drugs and/or their metabolites from biological samples of different sources. This can be used in pharmacokinetics, drug metabolism, biotransformation, and bioaccumulation analyses.
For full quantification, we use standard calibration curves and internal standards selected for the study. Our lab is currently non-GMP, but method validation can be performed following the FDA and EMA guidelines.
Includes method development and sample analyses for the detection and quantitation of other metabolites not covered in the broad screening panels. The lack of coverage could be due to many reasons. We can use our expertise to offer different strategies to overcome them. For instance:
- Lack of sensitivity. For low abundant metabolites, we can improve their detection by using special sample treatment protocols using our automated extraction system from Extrahera and analyzing the samples in the most sensitive triple-quadrupole from the core.
- Lack of specificity. Small molecules can be present in several isomeric species such as cis-trans or R/S. If necessary, we can use further develop our separation strategies (based on UPLC and SFC) to ensure that the measured form is the one of interest.
For full quantification, we use standard calibration curves and internal standards selected for the study. Our lab is currently non-GMP, but method validation can be performed following the FDA and EMA guidelines.
Equipment
- Waters Xevo TQ-S + Waters UPLC H-Class
- Waters Xevo TQ-S u + Waters UPLC Premier
- Waters Xevo TQ-XS + Waters UPLC Premier
- Waters Xevo TQ-XS + Waters SFC
- Waters Xevo-TQ Absolute + Waters UPLC i-Class
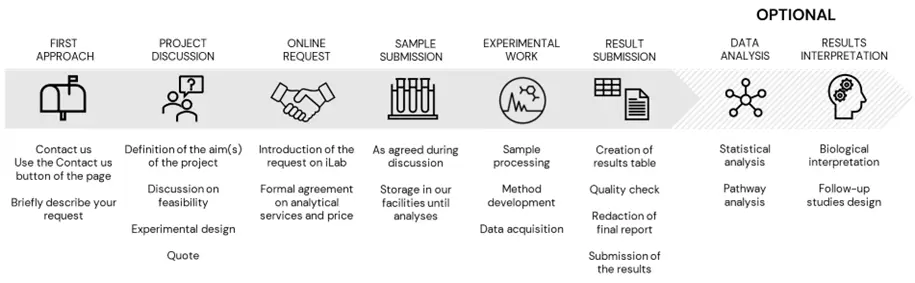
Our process
Both the analyses of a few metabolites or the profiling of hundreds of lipids require a proper experimental design to ensure that the results are of enough quality to answer your biological question.
We can assist in the planning, as well as in all the different steps of the process.
Our pricing model
Our services are billed according to a fee-for-service basis, in accordance with Karolinska Insitutet's policies for core facilities.
The user fees for Swedish academic institutions are based on cost for consumables, university OH, and part of the cost for staff, instrumentation and rent.
Fees for academics outside Sweden include full cost for staff, instrumentation and rent.
Commercial users pay full costs including staff, premises, consumables and an added 50% margin.
Please log in to iLab to see current pricing
External use of the core facility
KI–SMMS can offer services without a commissioned/contract research or collaborative research agreement, in accordance with the Swedish governmental ordinance (2022:1378) on fees for research infrastructure.
Find out more about external use of KI's core facilities
