Research Division of Chemistry I
We use mass spectrometry for studying various processes of biological and medical importance. The Division consists of the Molecular Biometry research group, proteomics core facility Proteomics Biomedicum and Chemical Proteomics Facility, which is part of the national Bio-MS facility.
Research interest
Proteomics
Proteomics is large-scale analysis of the expression levels of proteins and/or the occupancy their modifications. Proteomics can also quantitatively assess the stability of proteins against proteolysis or thermal unfolding.
Our goal is to perfect the proteomics analysis and expand its application to a broad range of biomedical problems. We envision time when proteomics analyses will be widely used in clinical practice. For helping KI researchers in addressing a broad range of biomedical problems with proteomics, we have established a proteomics core facility Proteomics Biomedicum (run by Dr. Hassan Gharibi).
Central dogma of proteomics
We formulated the “Central dogma of proteomics” which postulates that important biological processes almost inevitably involve changes in the expression levels, PTM occupancies or stability of the proteins involved in these processes.
We are also strong believers in Chemical proteomics, which studies by proteomics methods:
- The effects of small molecules, e.g. drugs, on cells and organisms.
- The properties of proteins using small molecules as probes.
For that we opened a Chemical proteomics core facility (run by Dr. Max Gaetani).
One of the predictions of the Central dogma of proteomics is that when a small-molecule drug kills mammalian cells (e.g. anticancer drug applied to cancer cells), the proteins whose expression changes the most are the drug target proteins. Empirical observations confirm this prediction, which forms the basis of the Functional Identification of Targets by Expression proteomics (FITExP). FITExP is one of the methods we are widely applying in Chemical proteomics projects. The other method is Thermal Proteome Profiling (TPP).
pl-Trap
In order to improve proteomics analysis, we have developed a novel tool for in-solution, on-line separation of proteins and peptides by their isoelectric point (pI). Under the commercial name “pI-Trap”, this tool is now being produced by Biomotif AB.
With the help of this tool we have verified the “pI≈7.4 hypothesis”. It postulates that in human blood, the pI region around 7.4 (corresponds to blood pH) is rich with abnormal proteoforms, which are suitable as biomarkers of e.g. neurodegenerative diseases.
We have developed a number of popular Open source tools for protein quantification – Quanti and DeMix Q for label-free analysis and a more general Diffacto. Prof. Lukas Käll was an important collaborator in the last two projects.
TopSpec - The most sophisticated platform for top-down analysis of intact proteins.
Project funded by the European Horizon 2020 research and innovation program under grant agreement no 829157. We envisioned a unique instrumental platform that combined novel fragmentation methods in MS/MS with ion mobility and a truly innovative ion deconvolution approach, as well as the best achievements in front-end separation merged with ultrahigh-resolution MS.
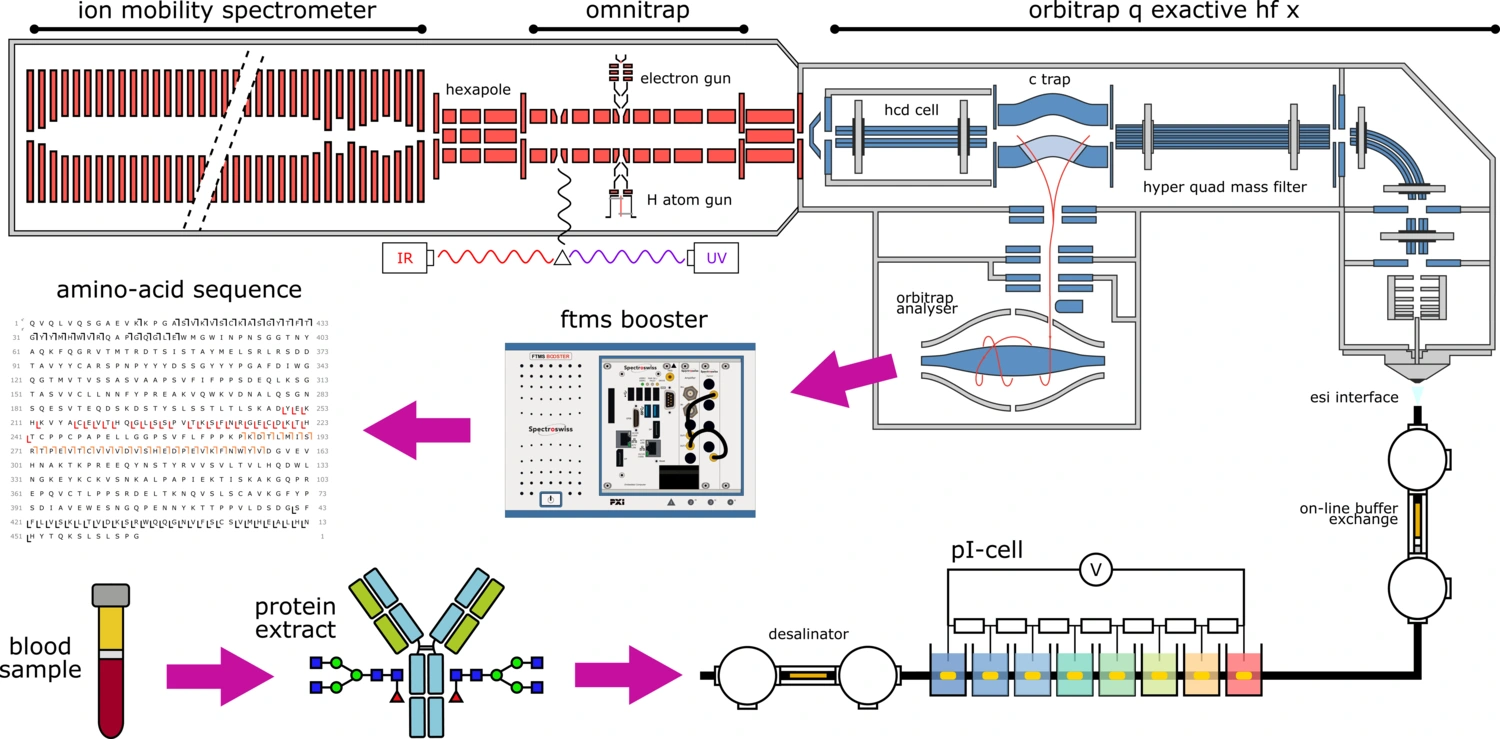
Learn more about the TopSpec project here:
Roman Zubarev (2022), "Next-generation precision antibody profiling; from science fiction to reality", Project Repository Journal, 15, pp.38-41. Available at https://www.europeandissemination.eu/article/next-generation-precision-antibody-profiling-from-science-fiction-to-reality/19532
Roman Zubarev (2023), "The TopSpec system: new levels in antibody characterisation by top-down mass spectrometry", Project Repository Journal, 16, pp.42-46. Available at https://www.europeandissemination.eu/article/the-topspec-system-new-levels-in-antibody-characterisation-by-top-down-mass-spectrometry/19878
Patents:
Biomotif:
METHOD FOR CAPILLARY ISOELECTRIC FOCUSING SEPARATION OF POLYPEPTIDES
Fasmatech:
SEGMENTED LINEAR ION TRAP FOR ENHANCED ION ACTIVATION AND STORAGE
ANALYSIS OF FRAGMENT IONS BY ION MOBILITY SEPARATION FOLLOWED BY MASS ANALYSIS
APPARATUS AND METHOD FOR ION ANYALYSIS USING MASS SPECTROMETRY
Peer-reviewed articles:
TDFragMapper: a visualization tool for evaluating experimental parameters in top-down proteomics, Dhenin J, Lima DB, Dupre M, Chamot-Rooke J. Bioinformatics. 2021;38(4):1136-8.
Software:
Free
Method development for top-down proteomics: FDFragMapper
TDFragMapper: a visualization tool for evaluating experimental parameters in top-down proteomics, Dhenin J, Lima DB, Dupre M, Chamot-Rooke J. Bioinformatics. 2021;38(4):1136-8.
New analysis tool for charge state determination in complex datasets: CHARDA
Adding colour to mass spectra: Charge Determination Analysis (CHARDA) assigns charge state to every ion peak, ChemRxiv, 17 Sept 2021
Conferences
We have initiated two series of annual international conferences:
- Uppsala Conference on Electron Capture/Transfer Dissociation and Related Phenomena (UppCon) – since 2003
Research groups and core facilities
- Molecular Biometry research group (Roman Zubarev group)
- Proteomics Biomedicum
- Chemical Proteomics core facility
Mass spectrometer

The division has a LC-MS/MS system based a Thermo Fusion Lumos Orbitrap mass spectrometer. MBB researchers with projects requiring ultrahigh sensitivity can get access to this instrument through our trained personnel. Our core facilities will be glad not only to provide service analyses in case of existing grants, but also to engage in collaboration by participating in joint grant applications.
For more information on the use of the instrument, please contact Roman Zubarev, Hassan Gharibi or Massimiliano Gaetani.
Contact
Roman Zubarev
ProfessorVisiting address
Division of Physiological Chemistry I
Biomedicum, 9A, floor 9
Solnavägen 9
171 65 Solna
