Pooled CRISPR Screening
CFG offers massively parallel genetic perturbations and genetic screening for unbiased discovery. Examples of our toolbox include genome-wide or custom CRISPR-based loss- and gain-of-function screens, small screens coupled with single cell transcriptomics as a readout, base-editing tiling mutagenesis screens and other modalities.
Service area
Pooled CRISPR screens are customizable, scalable, and cost-effective tools in both de novo discovery and functional exploration of phenotypes of interest. CFG has been specializing in applying and further developing this technology since 2017, incorporating in-house methods such as barcoded guide libraries.
CFG offers screening with Cas9, Cas12, and Cas13, as well as with fusions of these enzymes to transcriptional repressors (CRISPR-inhibition, CRISPR-i), activators (CRISPR-activation, CRISPR-a), and base-editors (scanning mutagenesis).
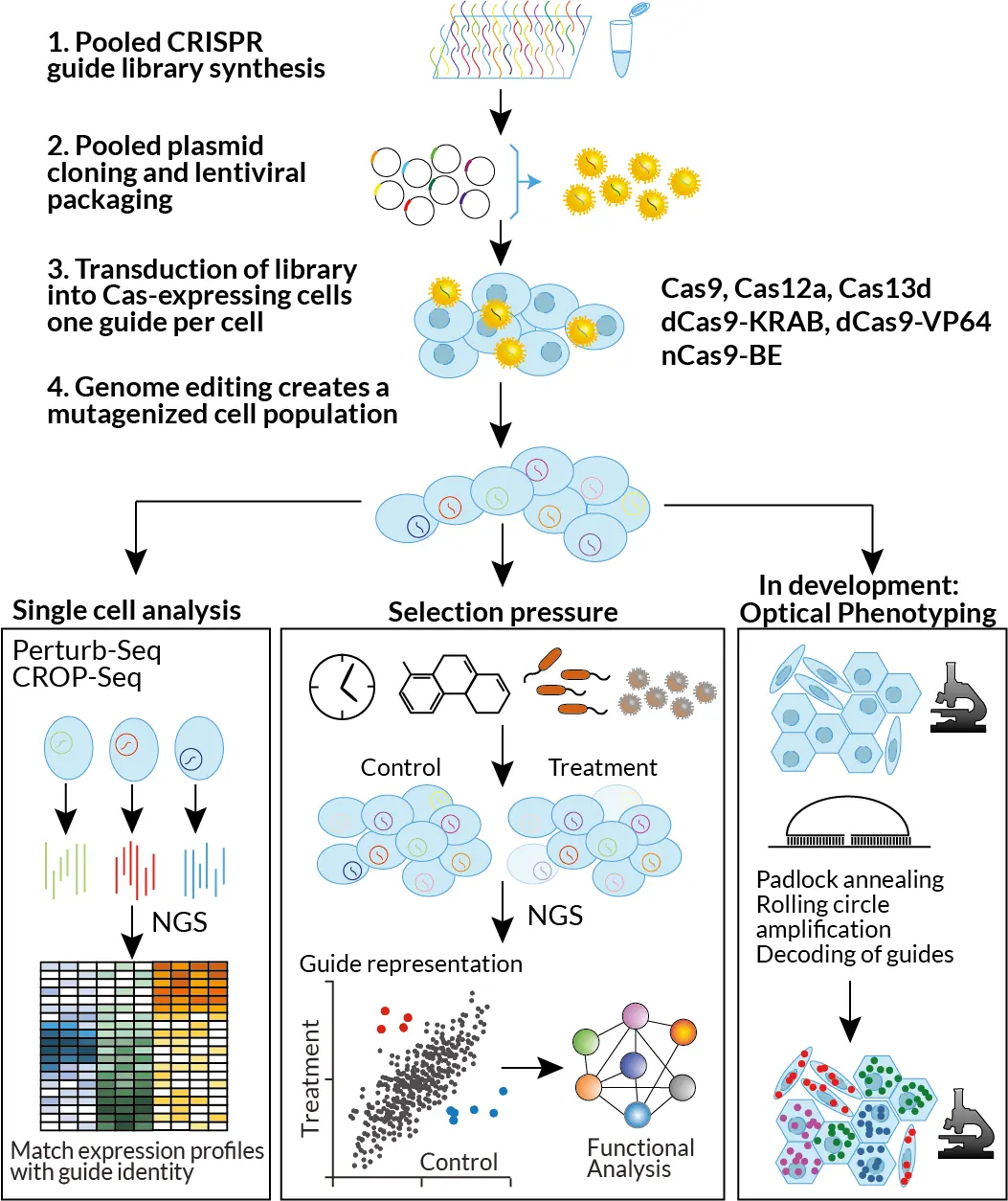
CFG can design and create any guide library requested. For screen deconvolution, we use either phenotypic selection (viability, FACS-based), single cell RNASeq (Perturb-Seq, CROP-Seq) or optical phenotyping (in development).
Our services
- Creation of stable Cas9/Cas12/Cas13 expressing cell lines (lentivirus, piggyBac)
- Creation of pooled, barcoded sgRNA libraries (100 – 150,000 guides), from design to packaged lentivirus
- Genome-wide/custom CRISPR-Cas9 KO, inhibition and activation screens for protein coding genes, miRNAs, lncRNAs, and regulatory genetic elements
- Pooled screens with single-cell RNASeq readout (in collaboration with NGI)
- Scanning mutagenesis with base-editors to characterize protein-protein or drug-target interactions
CFG Pooled genetic screening modalities
Examples screens
- Protein-coding genes: Genome-wide KO, inhibition, activation
- lncRNAs: Inhibition and activation of 20,000 lncRNAs
- Non-coding elements: CRISPR-inhibition on open chromatin regions
- RNA targeting: Cas13d screen for targetable loci in SARS-CoV2
- Synthetic interactions: Cas12a dual guide screen, with 112,000 guide combos
- 3D models: Genome-wide CRISPR-KO screen in pancreatic organoids
- Primary cells: miRNA-targeting screen in primary keratinocytes
- Perturb-Seq: Knock-out 50 genes in primary epithelial cells
- Tiling screens: 3’UTR tiling, CDS tiling
Creation of Cas expressing cell lines
Stable Cas-expressing cell lines are crucial for a successful screen. We use our own constructs expressing Cas-enzymes and a fluorescent marker, to create pools of cells stably expressing these enzymes, either by lentiviral or piggybac-based delivery. The fluorescent marker allows sorting of stable expressors, and regular monitoring of the cell line, which is critical due to silencing of integrated constructs.
Off-the-shelf and custom-made libraries
CFG clones all libraries in house to enable barcoding of guides. We have the following barcoded libraries ready for use:
- Human and mouse genome-wide KO library, four guides per gene (Brunello, Brie)
- Human genome-wide CRISPR-inhibition library, five guides per gene (Dolcetto)
- Human genome-wide CRISPR-activation library, four guides per gene (Calabrese)
- Human chromatin modifiers (around 1,200 genes), four guides per gene
For custom libraries, we work together with the client in designing a suitable library, order the oligo pools, clone, sequence verify and package into lentivirus.
Lentiviral library delivery
The lentiviral library is typically transduced into Cas-expressing cells at a low multiplicity of infection (MOI). This ensures that most cells receive a single integrated guide RNA. Following transduction, infected cells are selected using antibiotic resistance or fluorescence-based methods. This process generates a mutagenized cell population in which each cell harbors one genetic perturbation, marked by its unique guide RNA.
Screens with phenotypic separation
The mutagenized cell population is then subjected to a selection pressure, such as drug treatment, exposure to an infectious agent, or simply time (e.g., live/dead or proliferation/arrest screens). Cells that better withstand the selection pressure will become enriched, while those unable to cope will be depleted. The same pattern applies to the guide RNAs within these cells, reflecting the impact of the associated genetic perturbations.
Alternatively, phenotypic separation can be achieved through fluorescence-activated cell sorting (FACS). For instance, phenotypes such as changes in cell morphology or fluorescence intensity can be used to isolate subpopulations. In these cases, cells with guides that enhance the desired phenotype will become enriched, while those suppressing it will be depleted.
CFG handles all steps downstream of phenotypic selection, including genomic DNA isolation, amplification of guide cassettes, NGS library preparation, submission for NGS at NGI, and comprehensive data analysis.
Screens with single cell transcriptomic readout
For smaller screens (up to around 500 guides), single cell RNA-Seq can be used to read out both the guide and the transcriptome of single cells (Perturb-Seq, CROP-Seq). CFG has a seamless pipeline with single cell services offered by NGI. CFG supports all steps from library- and strategy design to a library-transduced cell population; helps with performing the desired treatment, and FACS sorts if needed. Viable cells are then delivered to NGI, who perform the single cell transcriptomics and data analysis.
Scanning mutagenesis screens with base editors
In a pooled scanning mutagenesis screen, a guide RNA library is designed to tile across one or more proteins of interest. Cas9 fused to base-editing enzymes introduces point mutations at the guide RNA binding sites, generating a diverse cell pool expressing a wide variety of mutated versions of the target protein(s). Mutations that confer a fitness advantage in the chosen assay can be pinpointed by identifying enriched guides. The specific mutations are then characterized through amplicon sequencing.
This versatile method can be applied to study drug-target interactions, protein-protein interactions, epitope mapping, and more.
Screens with optical phenotyping – in development
Due to the requirement for physical separation of phenotypes, complex or subtle phenotypes cannot be easily interrogated in conventional pooled screens. Screens with single cell transcriptomic readout overcome this limitation, but are often too expensive to perform at scale. We are thus implementing guide readout directly on a microscopy slide in collaboration with Mats Nilsson’s lab at SU and the SciLifeLab ISS unit.
With this method, cells can be phenotyped by microscopy, followed by determining the guide present in each cell by padlock-based rolling circle amplification and microscopic deconvolution of padlock barcodes by detecting rolling circle products with cycles of hybridization and stripping of fluorescently labelled deconvolution oligo libraries.
