Peter Swoboda
Cilia in the brain – and their connections to human brain disorders
Cilia are sensory or signalling structures projecting off cell surfaces like an antenna. In humans, many different cell types are ciliated, including neurons in the brain. Neuronal cilia as signalling hubs are involved in shaping and maintaining functional neuronal circuits. These circuits are crucial for orchestrating behavioural output. Ciliopathies are characterised by defective cilia and comprise various disease states, including brain phenotypes.
We have uncovered highly relevant connections between (i) cilia, ciliary genes and cilia-based signalling and (ii) (candidate genes for) different human brain conditions or disorders, like dyslexia or schizophrenia. The biological pathways behind these brain phenotypes are largely unknown. And our understanding of neuronal cilia is still rudimentary.
Fundamental questions about ciliary involvement in brain development, function and behavioural output in normal and disease states remain. What are the mechanisms by which cilia orchestrate cellular signalling in neuronal development? When and how does ciliary signalling control cell fate during expansion of the neural progenitor cell pool and their differentiation and organisation into neurons during brain development? Addressing these questions is crucial for understanding disease aetiology and for eventually developing treatment regimens for brain disorders.
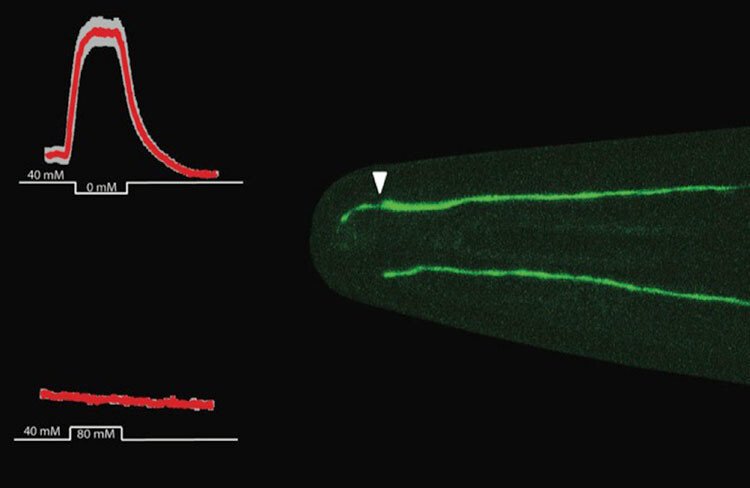
Our work lets us hypothesise that proper cilia function impacts differentiation of neural progenitor cells and early-stage neurons, including polarisation and neurite outgrowth, and thereby neuronal migration and circuit formation later on. Different non-lethal cilia malfunctions may thus cause different brain phenotypes (or disease states) depending on the affected neuron type and its location in the brain.
We address these hypotheses by:
- Determining in cultured human neurons and a whole-animal model, the worm C. elegans, causative connections between cilia formation, structure and function and aspects of neuronal development.
- Analysing transcriptomes of differentiating (human) neurons, including bioinformatics-based cross-correlations with ciliary gene lists and human candidate genes for various brain phenotypes.
- Analysing in the worm C. elegans behavioural output of disease-associated mutations in evolutionarily conserved (human and worm) ciliary genes connected to brain phenotypes.
Our goal is to create proper spatiotemporal information about ciliary localisation and function of proteins encoded by disease genes to better understand human brain development in the context of neural progenitor cells, and differentiating and mature functional neurons.
Group members
Peter Swoboda
Group leaderAndrea Coschiera
Phd StudentHaonan Li
Phd StudentMasahito Yoshihara
Affiliated to ResearchMariangela Pucci
Guest Researcher (University of Teramo, Italy)Selected publications
- The C. elegans regulatory factor X (RFX) DAF-19M module: A shift from general ciliogenesis to cell-specific ciliary and behavioral specialization.
Ahn S, Yang H, Son S, Lee HS, Park D, Yim H, Choi HJ, Swoboda P, Lee J
Cell Rep 2022 Apr;39(2):110661
- Differentiation of ciliated human midbrain-derived LUHMES neurons.
Lauter G, Coschiera A, Yoshihara M, Sugiaman-Trapman D, Ezer S, Sethurathinam S, et al.,
J Cell Sci 2020 Nov;133(21):jcs249789
- Lauter G, Swoboda P, Tapia-Páez I. (2018), ’Cilia in brain development and disease.’ Cilia: Development and Disease. 2018 May 31 (ed. P. Goggolidou, CRC Press, Taylor and Francis Publishers, Boca Raton, Florida, USA); ch. 1:pp. 1-35.
- DAF-16/FOXO and HLH-30/TFEB function as combinatorial transcription factors to promote stress resistance and longevity.
Lin XX, Sen I, Janssens GE, Zhou X, Fonslow BR, Edgar D, Stroustrup N, Swoboda P, Yates JR, Ruvkun G, Riedel CG
Nat Commun 2018 Oct;9(1):4400
- Characterization of the human RFX transcription factor family by regulatory and target gene analysis.
Sugiaman-Trapman D, Vitezic M, Jouhilahti EM, Mathelier A, Lauter G, Misra S, et al.,
BMC Genomics 2018 03;19(1):181
- Ciliary dyslexia candidate genes DYX1C1 and DCDC2 are regulated by Regulatory Factor X (RFX) transcription factors through X-box promoter motifs.
Tammimies K, Bieder A, Lauter G, Sugiaman-Trapman D, Torchet R, Hokkanen ME, et al.,
FASEB J 2016 10;30(10):3578-3587
- Neuropeptidergic Signaling and Active Feeding State Inhibit Nociception in Caenorhabditis elegans.
Ezcurra M, Walker DS, Beets I, Swoboda P, Schafer WR
J Neurosci 2016 Mar;36(11):3157-69
Research networks
- KI Neurosciences network (StratNeuro)
- European C. elegans researcher network (EU COST Action Network BM1408)
- Swedish-Korean research cooperation network (VR and STINT, Korean NRF)
